Search Count: 16
 |
Organism: Ideonella sakaiensis (strain nbrc 110686 / tistr 2288 / 201-f6)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: BEZ, CA |
 |
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: CA |
 |
Organism: Ideonella sakaiensis (strain 201-f6)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-09-30 Classification: STRUCTURAL GENOMICS Ligands: BEZ, CA |
 |
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: CA, SO4 |
 |
Structural And Functional Analysis Of The Hordeum Vulgare L. Hvgr-Rbp1 Protein, A Glycine-Rich Rna Binding Protein Implicated In The Regulation Of Barley Leaf Senescence And Environmental Adaptation
Organism: Hordeum vulgare
Method: SOLUTION NMR Release Date: 2014-12-17 Classification: RNA BINDING PROTEIN |
 |
Solution Structure And Molecular Determinants Of Hemoglobin Binding Of The First Neat Domain Of Isdb In Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2014-07-02 Classification: PROTEIN BINDING |
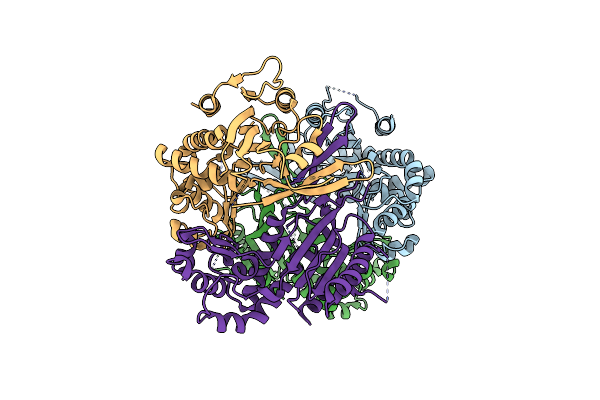 |
The Structure Of The Crispr-Associated Protein, Csa2, From Sulfolobus Solfataricus
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-04-20 Classification: RNA BINDING PROTEIN |
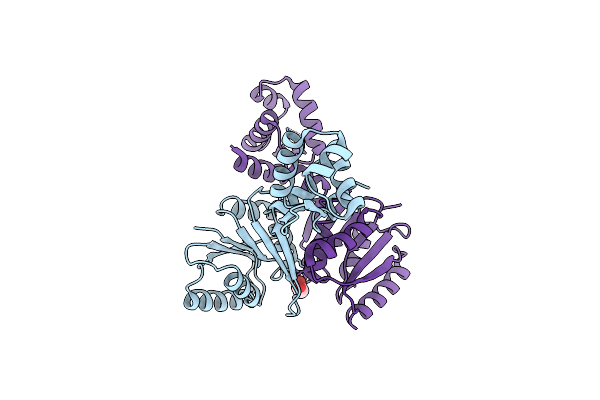 |
The Structure Of The Crispr-Associated Protein, Csa3, From Sulfolobus Solfataricus At 1.8 Angstrom Resolution.
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-09-22 Classification: ANTIVIRAL PROTEIN Ligands: PEG |
 |
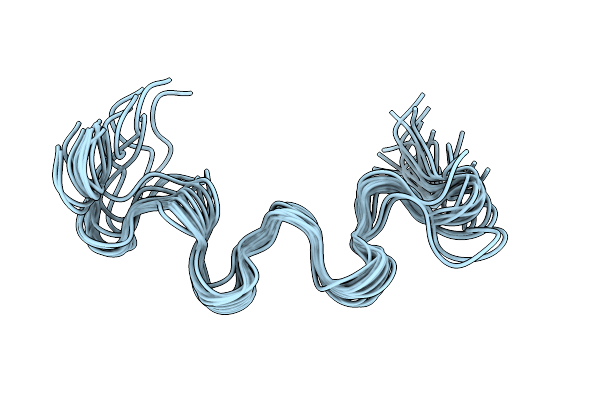
Tryptophan Repressor With L75F Mutation In Its Apo Form (No L- Tryptophan Bound)
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2010-07-07 Classification: TRANSCRIPTION |
 |
Nmr Structure Of The Octarepeat Region Of Prion Protein Bound To Pentosan Polysulfate
Organism: Mesocricetus auratus
Method: SOLUTION NMR Release Date: 2009-11-10 Classification: MEMBRANE PROTEIN |
 |
Solution Nmr Structure Of The Apo-Nosl Protein From Achromobacter Cycloclastes
Organism: Achromobacter cycloclastes
Method: SOLUTION NMR Release Date: 2006-10-24 Classification: METAL TRANSPORT |
 |
Solution Nmr Structure Of The Apo-Nosl Protein From Achromobacter Cycloclastes
Organism: Achromobacter cycloclastes
Method: SOLUTION NMR Release Date: 2006-10-24 Classification: METAL TRANSPORT |
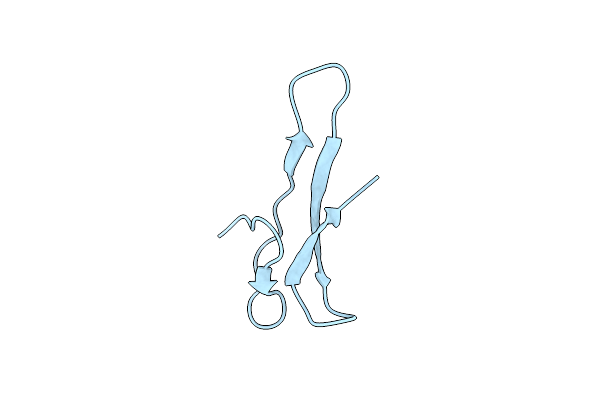 |
Nmr Structure Of The Carboxyl-Terminal Cysteine Domain Of The Vhv1.1 Polydnaviral Gene Product
Organism: Campoletis sonorensis ichnovirus
Method: SOLUTION NMR Release Date: 2004-10-05 Classification: VIRAL PROTEIN |
 |
3D Solution Structure Of The C-Terminal Cysteine-Rich Domain Of The Vhv1.1 Polydnaviral Gene Product
Organism: Campoletis sonorensis ichnovirus
Method: SOLUTION NMR Release Date: 2004-10-05 Classification: VIRAL PROTEIN |
 |
Solution Nmr Structure Of Linked Cell Attachment Modules Of Mouse Fibronectin Containing The Rgd And Synergy Regions, 20 Structures
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 1998-04-29 Classification: CELL ADHESION PROTEIN |
 |
Solution Nmr Structure Of Linked Cell Attachment Modules Of Mouse Fibronectin Containing The Rgd And Synergy Regions, 10 Structures
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 1998-04-29 Classification: CELL ADHESION PROTEIN |

