Search Count: 14
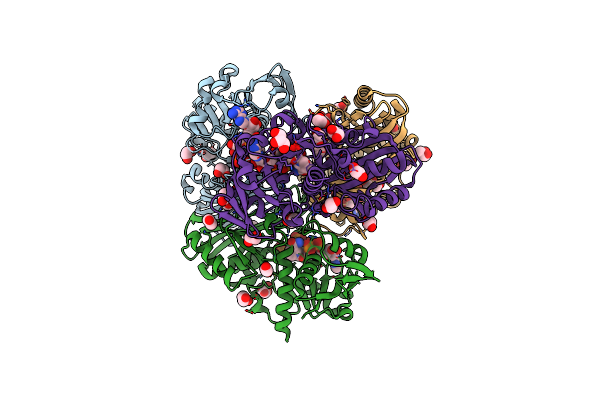 |
A Fungal Glyceraldehyde-3-Phosphate Dehydrogenase With Self-Resistance To Inhibitor Heptelidic Acid
Organism: Hypocrea virens (strain gv29-8 / fgsc 10586)
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: EDO, GOL, NAD, PO4, PEG |
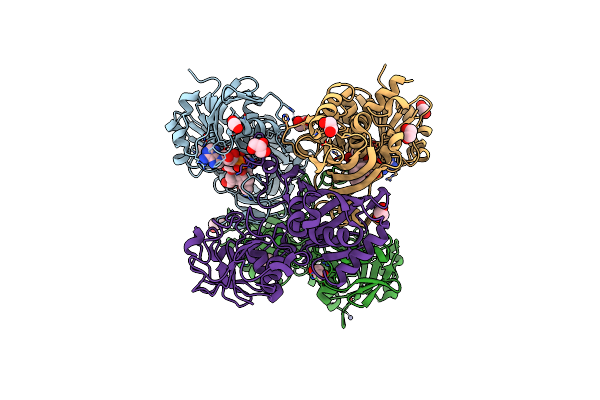 |
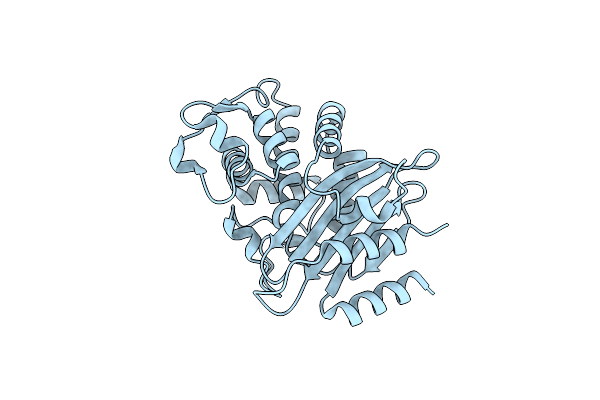
Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) With Inhibitor Heptelidic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: EDO, PEG, ZN, F4F, NAD, GOL |
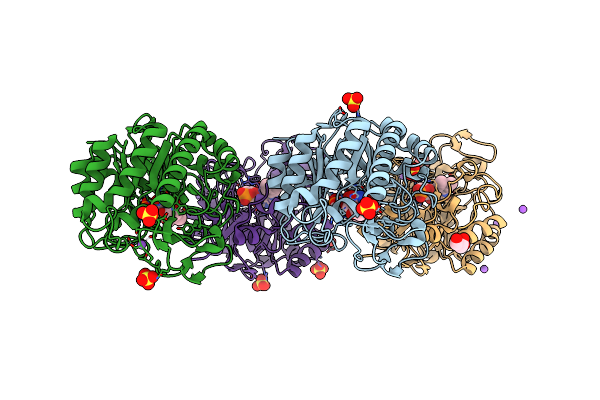 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: FMN, ACT, SO4, NA, GOL |
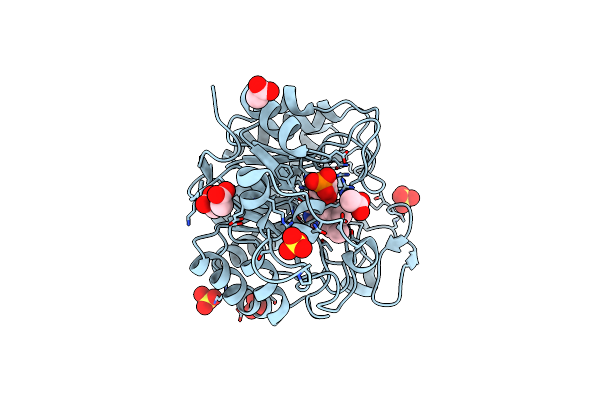 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: FMN, SO4, ACT, NA, CL |
 |
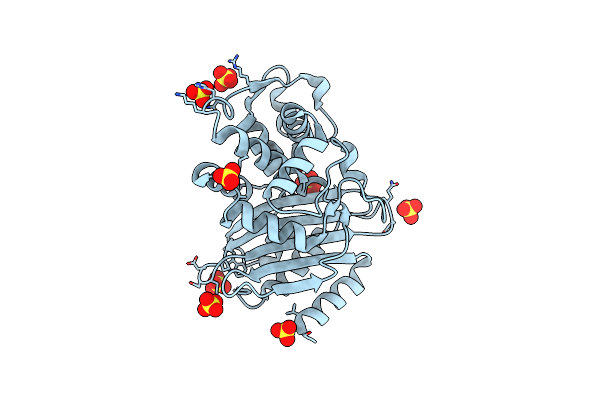
Substrate Binding Induces Conformational Changes In A Class A Beta Lactamase That Primes It For Catalysis
Organism: Escherichia coli
Method: NEUTRON DIFFRACTION Resolution:1.75 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: DOD |
 |
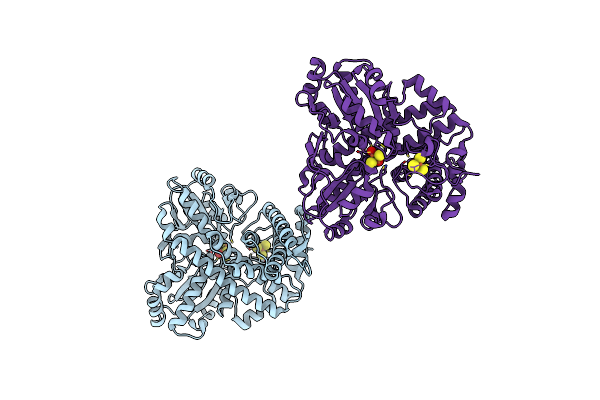
Conformational Changes In A Class A Beta Lactamase That Prime It For Catalysis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: CE3, SO4 |
 |
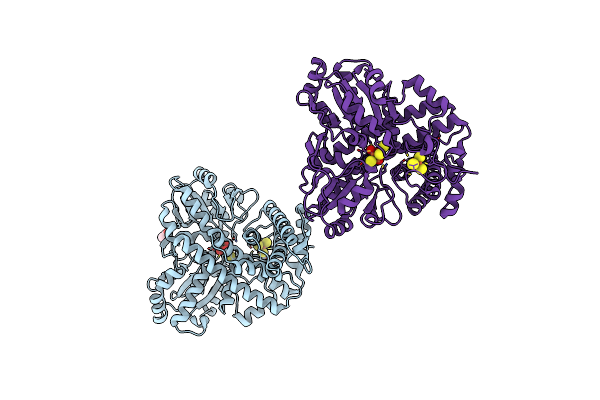
Conformational Changes In A Class A Beta Lactamase That Prime It For Catalysis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: SO4 |
 |
Hybrid Cluster Protein From Desulfovibrio Desulfuricans Atcc 27774 X-Ray Structure At 2.6A Resolution Using Synchrotron Radiation At A Wavelength Of 1.722A
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2002-04-11 Classification: HYBRID CLUSTER Ligands: FSO, SF4 |
 |
Hybrid Cluster Protein From Desulfovibrio Desulfuricans X-Ray Structure At 1.25A Resolution Using Synchrotron Radiation At A Wavelength Of 0.933A
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Release Date: 2002-04-11 Classification: HYBRID CLUSTER PROTEIN Ligands: SF4, FSO, ACT |
 |
Hybrid Cluster Protein From Desulfovibrio Vulgaris. X-Ray Structure At 1.25A Resolution Using Synchrotron Radiation.
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Release Date: 2002-04-11 Classification: OXIDOREDUCTASE Ligands: SF4, FSO |
 |
Xenon Bound In Hydrophobic Channel Of Hybrid Cluster Protein From Desulfovibrio Vulgaris
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2001-10-25 Classification: OXIDOREDUCTASE Ligands: FSO, SF4, XE, EDO, ACY, GOL, TRS |
 |
Low Temperature Structure Of Hybrid Cluster Protein From Desulfovibrio Vulgaris To 1.6A
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2000-12-11 Classification: OXIDOREDUCTASE Ligands: SF4, FSO |
 |
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2000-06-21 Classification: OXIDOREDUCTASE Ligands: FSO, SF4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1997-07-07 Classification: LYASE Ligands: ZN |

