Search Count: 35
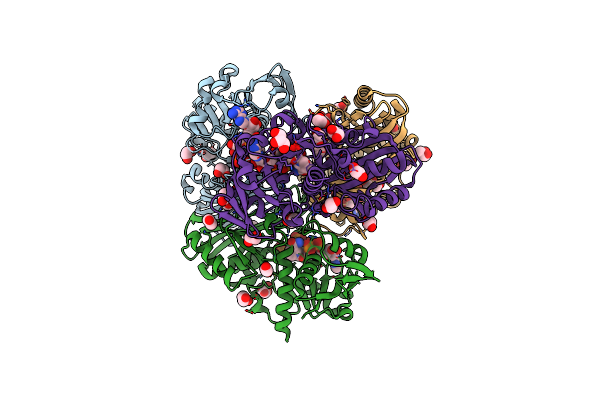 |
A Fungal Glyceraldehyde-3-Phosphate Dehydrogenase With Self-Resistance To Inhibitor Heptelidic Acid
Organism: Hypocrea virens (strain gv29-8 / fgsc 10586)
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: EDO, GOL, NAD, PO4, PEG |
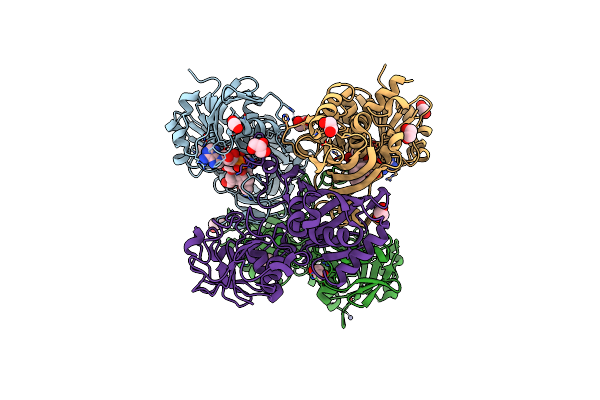 |
Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) With Inhibitor Heptelidic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: EDO, PEG, ZN, F4F, NAD, GOL |
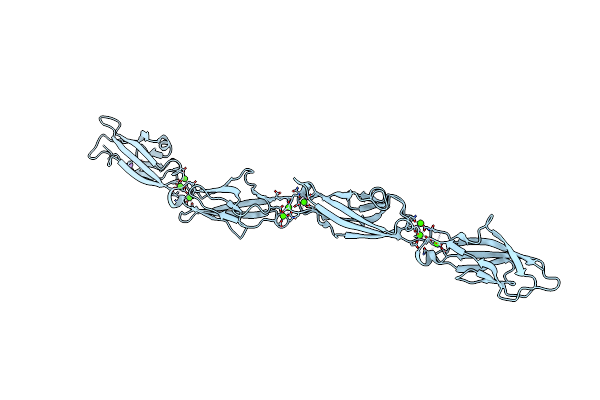 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-12-30 Classification: CELL ADHESION Ligands: CA, NA |
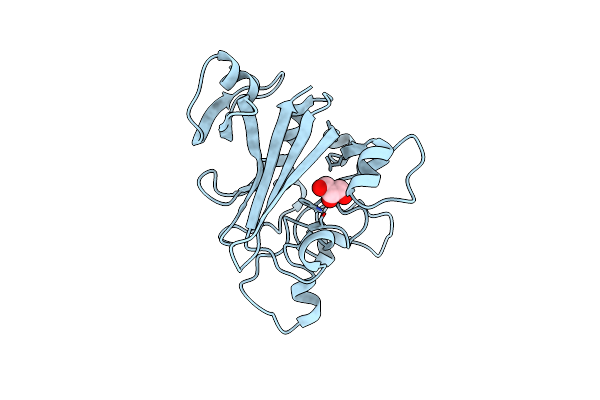 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2019-10-09 Classification: SIGNALING PROTEIN Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-10-09 Classification: SIGNALING PROTEIN Ligands: PLM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2019-10-09 Classification: SIGNALING PROTEIN Ligands: MYR |
 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: FMN, ACT, SO4, NA, GOL |
 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: FMN, SO4, ACT, NA, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2019-06-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2019-06-12 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2019-06-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2019-06-12 Classification: DE NOVO PROTEIN |
 |
Substrate Binding Induces Conformational Changes In A Class A Beta Lactamase That Primes It For Catalysis
Organism: Escherichia coli
Method: NEUTRON DIFFRACTION Resolution:1.75 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: DOD |
 |
Conformational Changes In A Class A Beta Lactamase That Prime It For Catalysis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: CE3, SO4 |
 |
Conformational Changes In A Class A Beta Lactamase That Prime It For Catalysis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2016-11-02 Classification: CELL ADHESION Ligands: CA |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2016-11-02 Classification: CELL ADHESION Ligands: CA, NA |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2016-09-21 Classification: UNKNOWN FUNCTION Ligands: ZN, GOL, SO4 |
 |
The Discovery Of Novel, Potent And Highly Selective Inhibitors Of Inducible Nitric Oxide Synthase (Inos)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-10-08 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, YWO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2013-12-18 Classification: MEMBRANE PROTEIN Ligands: FMT, CA |

