Search Count: 47
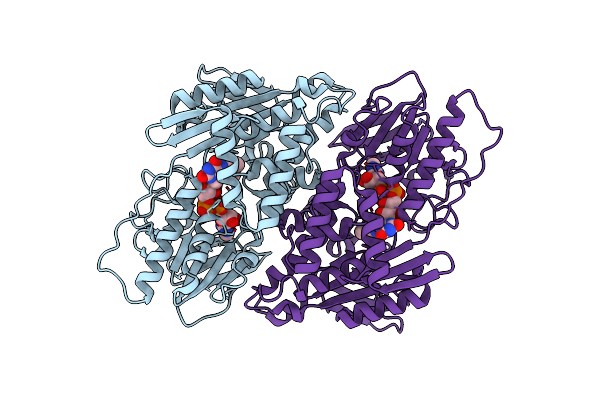 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN Ligands: FAD |
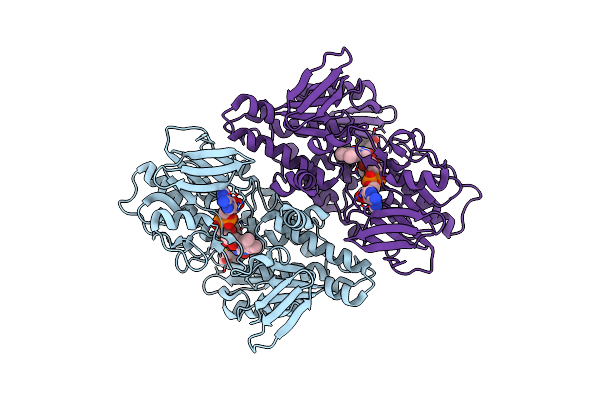 |
Structure Of Mycobacterial Ndh2 (Type Ii Nadh:Quinone Oxidoreductase) With 2-Mercapto-Quinazolinone Inhibitor.
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN Ligands: FAD, A1BNR |
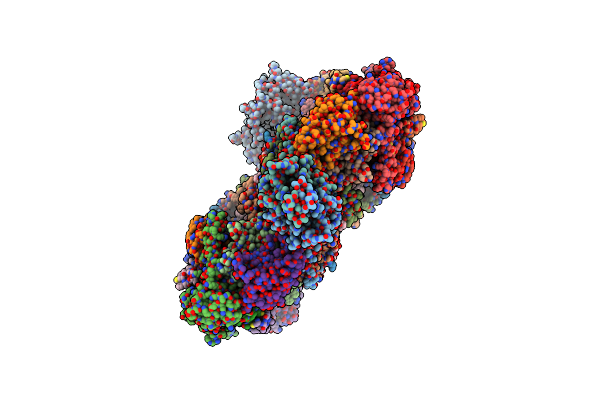 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: CU, HEA, CDL, HEM, MQ9, 9Y0, PLM, 9XX, HEC, 9YF, FES |
 |
The 2.19-Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Complex Minus Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.67 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, OH, MG, VK3, F3S |
 |
The Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Full Complex Focused Refinement Of Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.52 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: ELECTRON TRANSPORT Ligands: O, MG, FCO, 3NI, VK3, F3S |
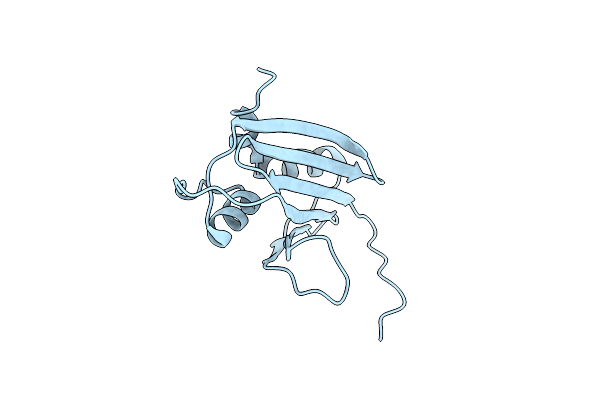 |
Structure Of 2A Protein From Theilers Murine Encephalomyelitis Virus (Tmev)
Organism: Theiler's murine encephalomyeltits virus gdvii
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2021-12-08 Classification: VIRAL PROTEIN Ligands: BR |
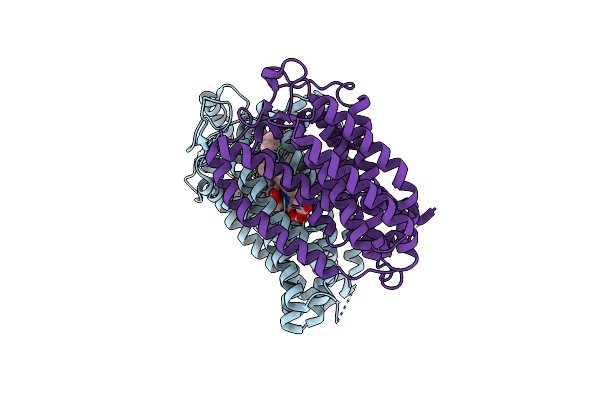 |
Cryo-Em Structure Of The Cytochrome Bd Oxidase From M. Tuberculosis At 2.5 A Resolution
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2021-09-22 Classification: MEMBRANE PROTEIN Ligands: OXY, MQ9, HDD, HEB |
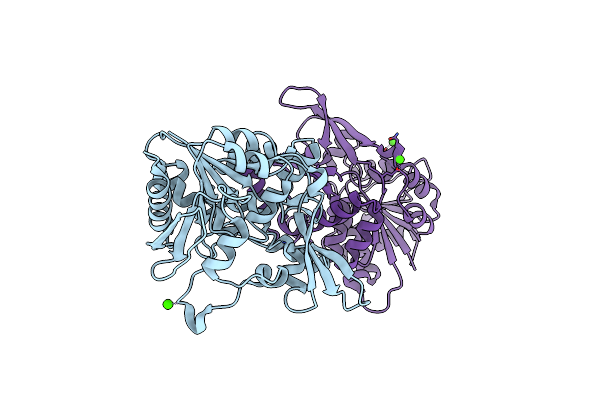 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: CA |
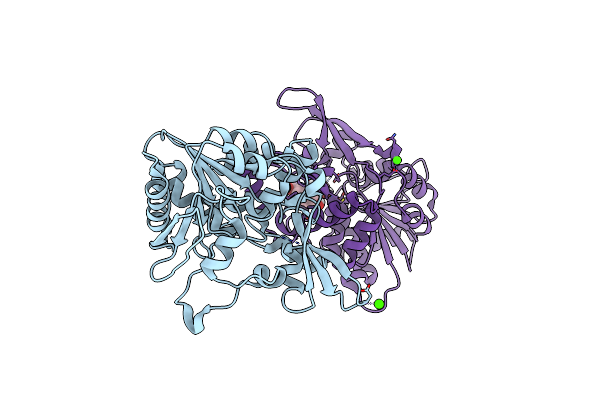 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FO1, CA |
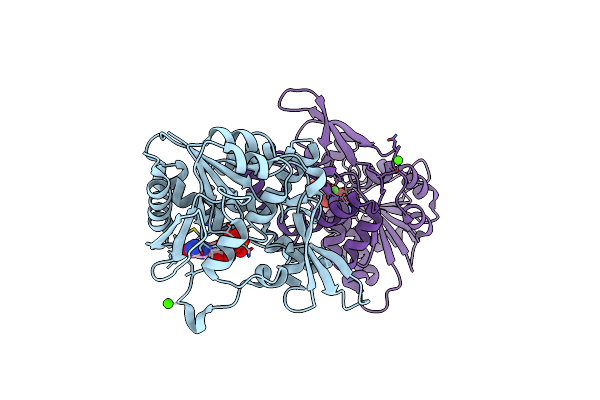 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, GOL, CA |
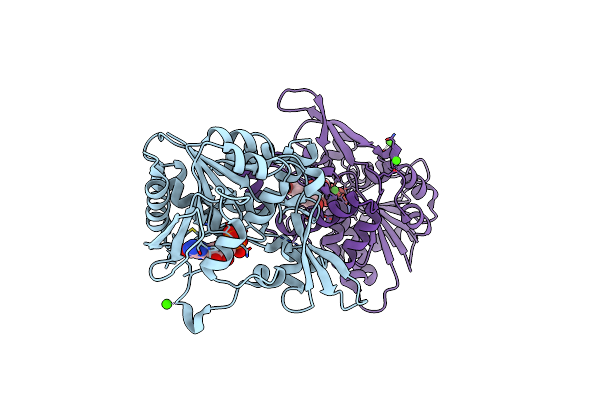 |
The Crystal Structure Of Fbia From Mycobacterium Smegmatis, Gdp And Fo Bound Form
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, FO1, CA |
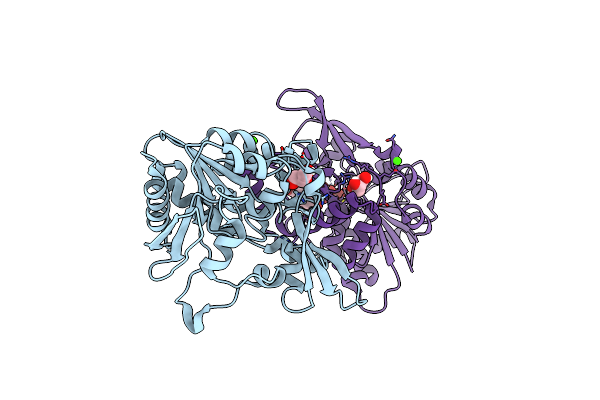 |
The Crystal Structure Of Fbia From Mycobacterium Smegmatis, Dehydro-F420-0 Bound Form
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, QMD, CA |
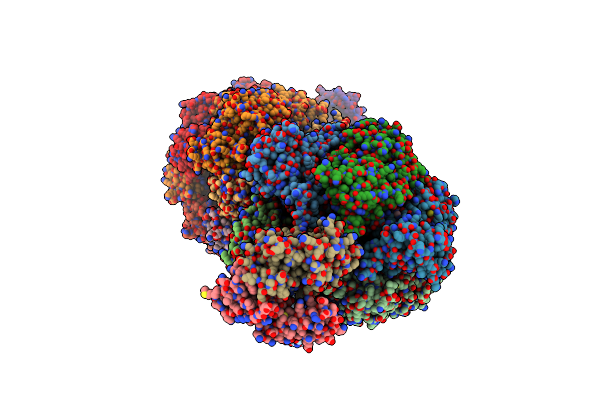 |
Organism: Fusobacterium nucleatum subsp. nucleatum atcc 25586
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2019-07-10 Classification: HYDROLASE Ligands: ATP, MG, ADP |
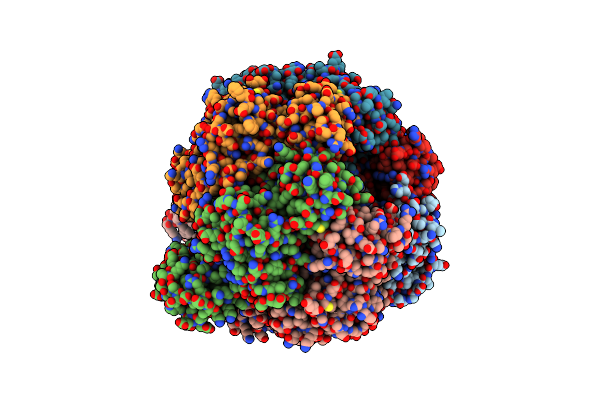 |
Organism: Mycolicibacterium smegmatis mc2 155, Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: ADP, MG, PO4 |
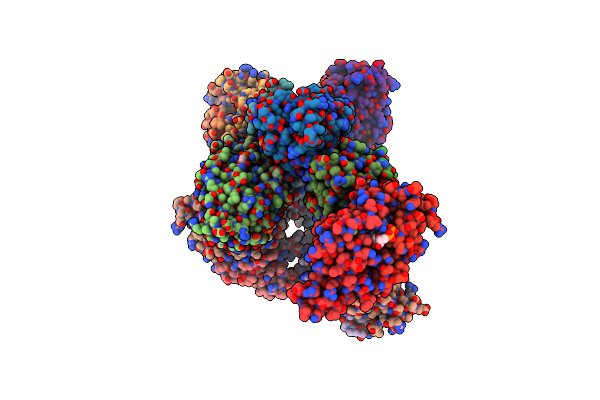 |
Structure Of The Fad Binding Protein Msmeg_5243 From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2018-12-26 Classification: FAD-BINDING PROTEIN Ligands: FAD, CL |
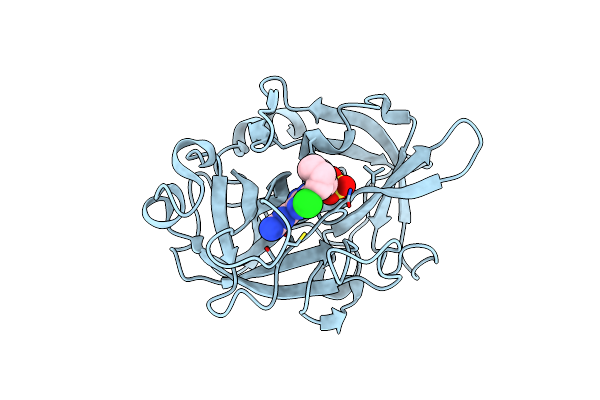 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-12-19 Classification: HYDROLASE Ligands: HMX, SO4 |
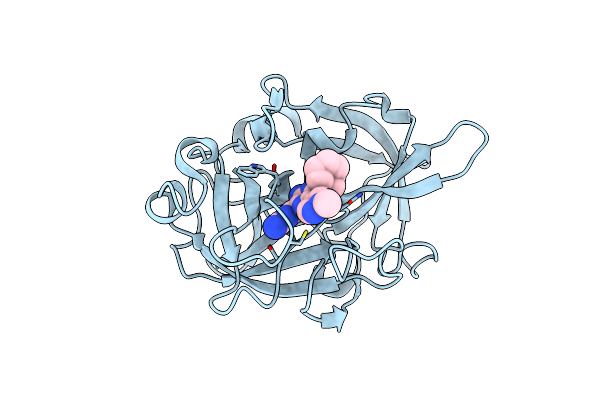 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-12-19 Classification: HYDROLASE Ligands: 27I |
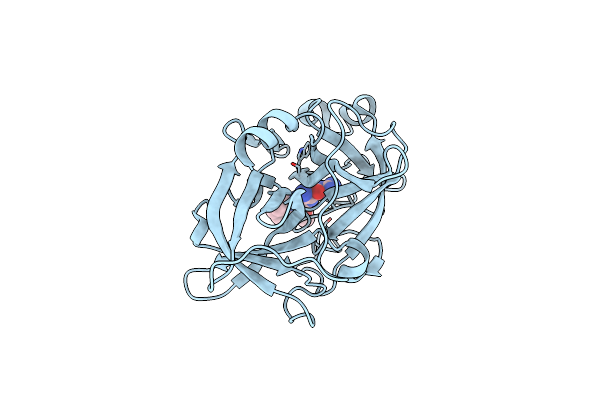 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2018-12-19 Classification: HYDROLASE Ligands: 50I |

