Search Count: 18
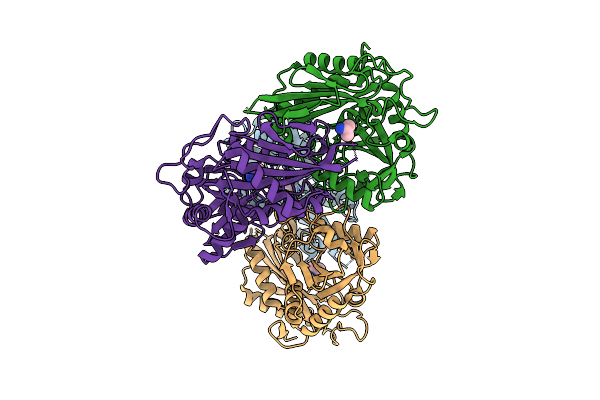 |
Structure Of Tf9 Mutant Of Aminotransferase From Mycolicibacterium Neoaurum In Complex With Llp And G4O
Organism: Mycolicibacterium neoaurum vkm ac-1815d
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: G4O |
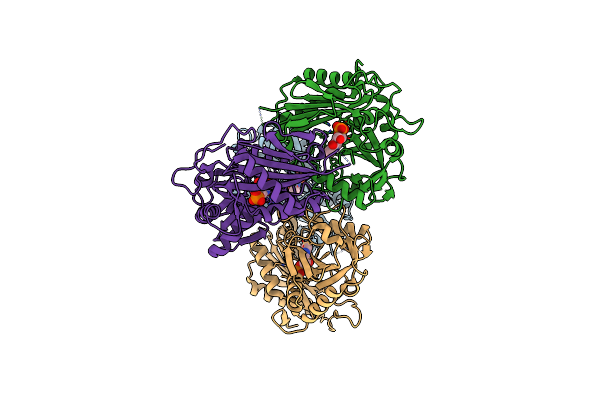 |
Structure Of Aminotransferase From Mycolicibacterium Neoaurum In Complex With Plp
Organism: Mycolicibacterium neoaurum
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: HYDROLASE Ligands: GOL, SO4 |
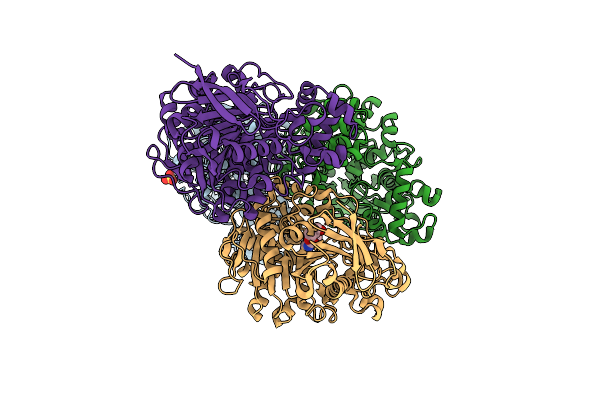 |
Crystal Structure Of A Novel Pu Plastic Degradation Enzyme With Ligand From Thermaerobacter Marianensis
Organism: Thermaerobacter marianensis (strain atcc 700841 / dsm 12885 / jcm 10246 / 7p75a)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: SO4, A1D5F |
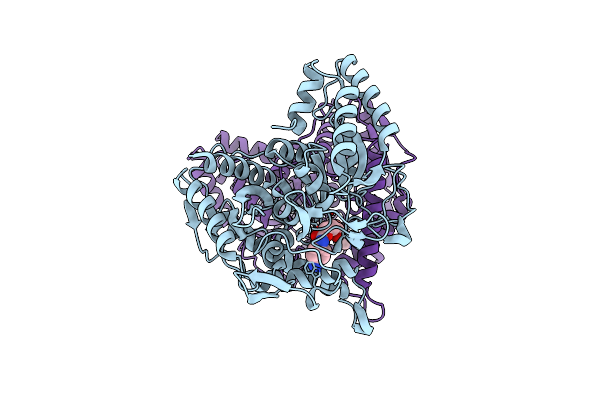 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 From Uncultured Bacterium In Complex With Ligand
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: A1D5H |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 Mutant From Uncultured Bacterium In Complex With Ligand
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: A1D5H, GOL |
 |
Crystal Structure Of Glucose 1-Dehydrogenase Mutant1 From Limosilactobacillus Fermentum
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2024-10-30 Classification: HYDROLASE |
 |
Crystal Structure Of Glucose 1-Dehydrogenase Mutant2 From Limosilactobacillus Fermentum
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-10-30 Classification: HYDROLASE |
 |
Crystal Structure Of Glucose 1-Dehydrogenase From Limosilactobacillus Fermentum
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-10-30 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of A Short-Chain Dehydrogenase From Lactobacillus Fermentum With Nadph
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-30 Classification: HYDROLASE Ligands: NAP |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 From Uncultured Bacterium
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: GOL, SO4, PMS |
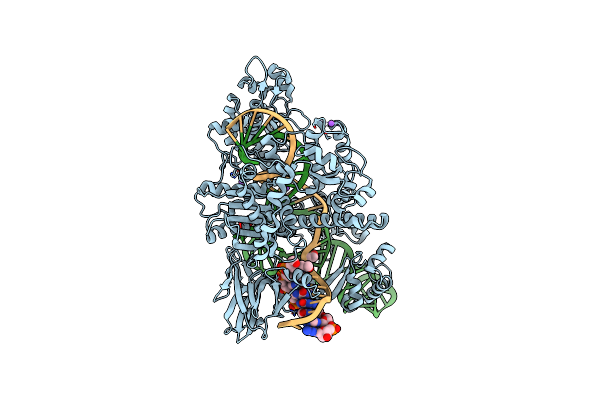 |
Crystal Structure Of Staphylococcus Aureus Cas9 In Complex With Sgrna And Target Dna (Ttgggt Pam)
Organism: Staphylococcus aureus subsp. aureus, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-09-02 Classification: HYDROLASE/RNA/DNA |
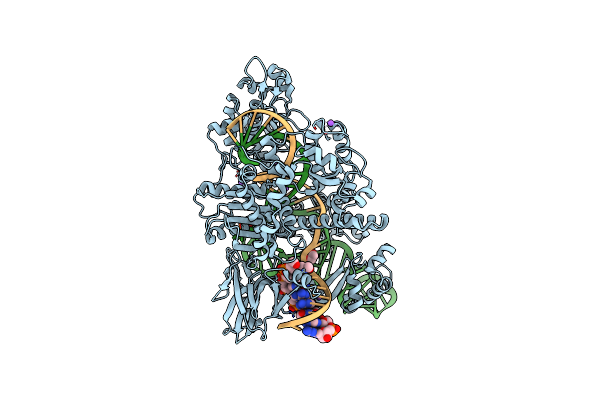 |
Crystal Structure Of Staphylococcus Aureus Cas9 In Complex With Sgrna And Target Dna (Ttgaat Pam)
Organism: Staphylococcus aureus subsp. aureus, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-09-02 Classification: HYDROLASE/RNA/DNA |
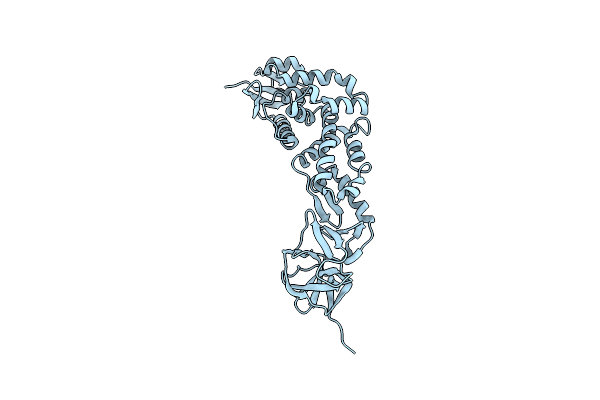 |
Organism: Avian infectious bronchitis virus (strain m41)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-05-25 Classification: HYDROLASE |
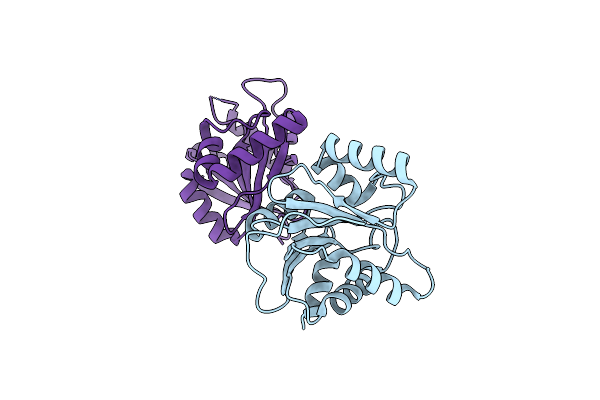 |
Organism: Avian infectious bronchitis virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-01-13 Classification: HYDROLASE |
 |
Organism: Avian infectious bronchitis virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-01-13 Classification: HYDROLASE Ligands: APR |
 |
Organism: Human coronavirus 229e
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-01-13 Classification: HYDROLASE |
 |
Organism: Human coronavirus 229e
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2009-01-13 Classification: HYDROLASE Ligands: APR |

