Search Count: 15
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: EDO |
 |
Crystal Structure Of Lbma Ox-Acp Didomain In Complex With Nadp And Ethyl Glycinate From The Lobatamide Pks (Gynuella Sunshinyii)
Organism: Gynuella sunshinyii
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2023-05-31 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, NAP, OXY, GEE, GOL, PO4 |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: PNS |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: PNS |
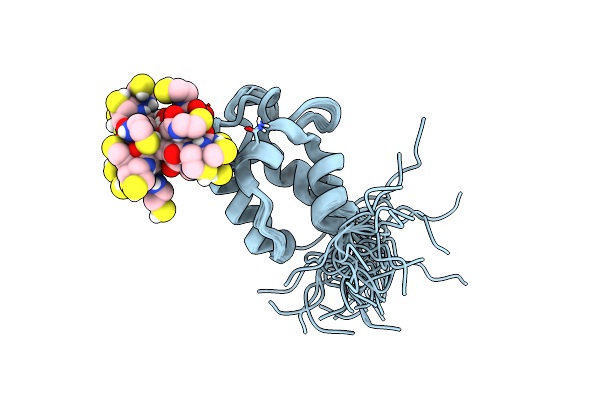 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: PNS |
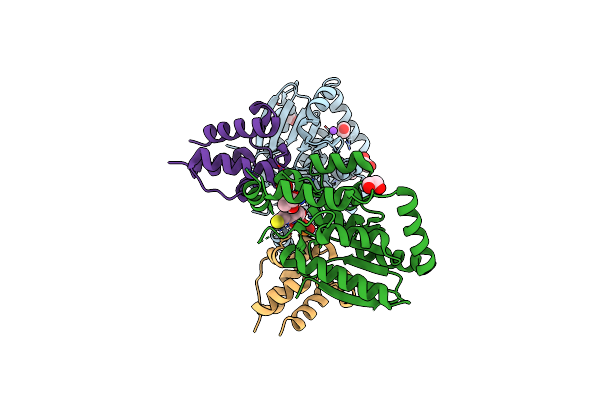 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-15 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, CL, NA, PNS |
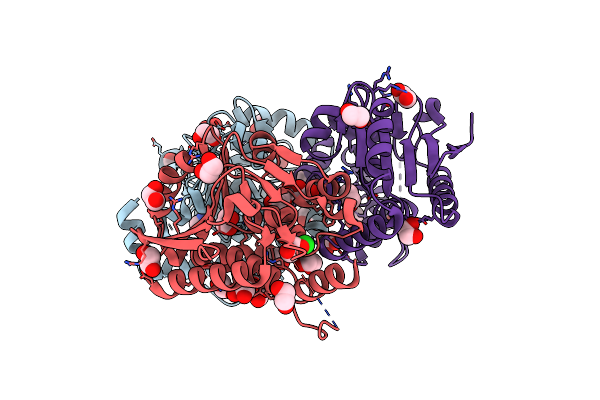 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-03-15 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, IMD, CL |
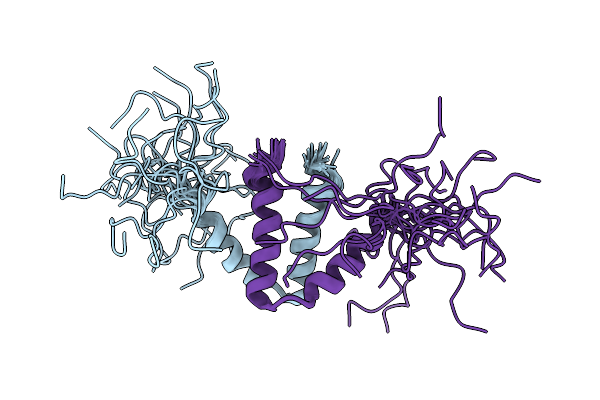 |
Organism: Burkholderia ambifaria ammd
Method: SOLUTION NMR Release Date: 2020-08-12 Classification: PROTEIN BINDING |
 |
Organism: Burkholderia ambifaria ammd
Method: SOLUTION NMR Release Date: 2020-08-12 Classification: PROTEIN BINDING |
 |
Organism: Burkholderia ambifaria ammd
Method: SOLUTION NMR Release Date: 2020-08-12 Classification: PROTEIN BINDING |
 |
Crystal Structure Of A Dual Function Amine Oxidase/Cyclase In Complex With Substrate Analogues
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, ACT, CJ8, CA, GOL, CJE |
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, CA, CWH, GOL, ACT |
 |
Organism: Streptomyces rochei subsp. volubilis
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, CA, CWH, GOL |
 |
Organism: Streptomyces rochei subsp. volubilis
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, OXY, NA |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2016-03-23 Classification: PROTEIN BINDING |

