Search Count: 21
 |
Solution Structure Of The Hemoglobin Receptor Hbpa From Corynebacterium Diphtheriae
Organism: Corynebacterium diphtheriae nctc 13129
Method: SOLUTION NMR Release Date: 2025-01-08 Classification: PROTEIN BINDING |
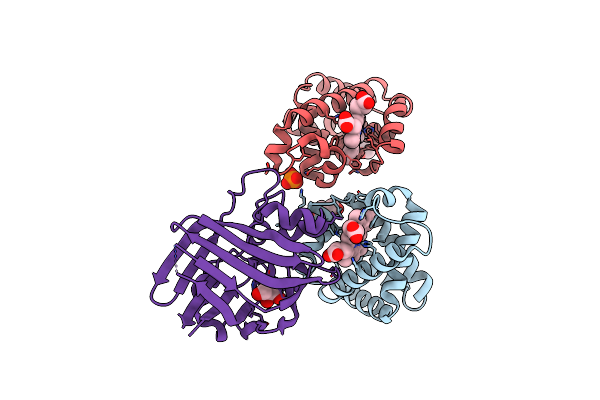 |
Crystal Structure Of Human Hemoglobin In Complex With The Hbpa Receptor From Corynebacterium Diphtheriae
Organism: Corynebacterium diphtheriae nctc 13129, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-01-08 Classification: PROTEIN BINDING Ligands: HEM, GOL, PO4 |
 |
Crystal Structure Of Mevalonate 3,5-Bisphosphate Decarboxylase From Picrophilus Torridus
Organism: Picrophilus torridus dsm 9790
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-12-22 Classification: LYASE Ligands: OLA |
 |
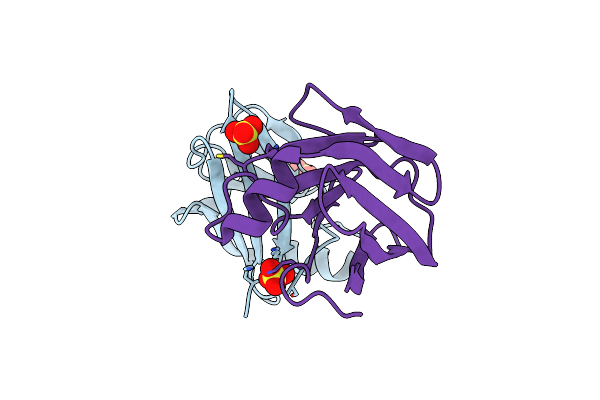
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-05-26 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
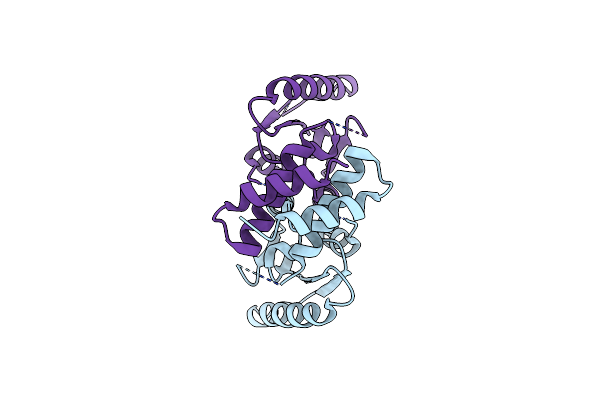
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-10-24 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
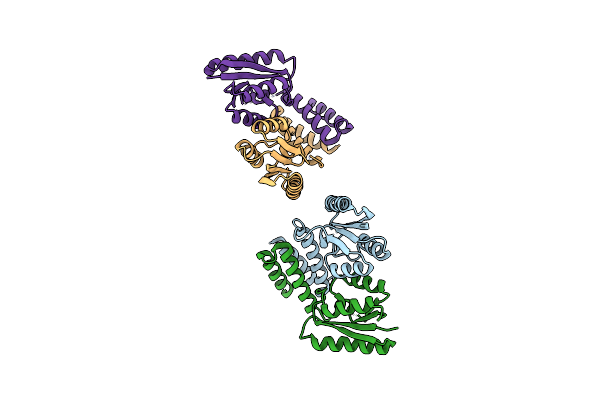
Crystal Structure Of The C-Terminal Catalytic Domain Of Is1535 Tnpa, An Is607-Like Serine Recombinase
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2018-07-18 Classification: HYDROLASE |
 |
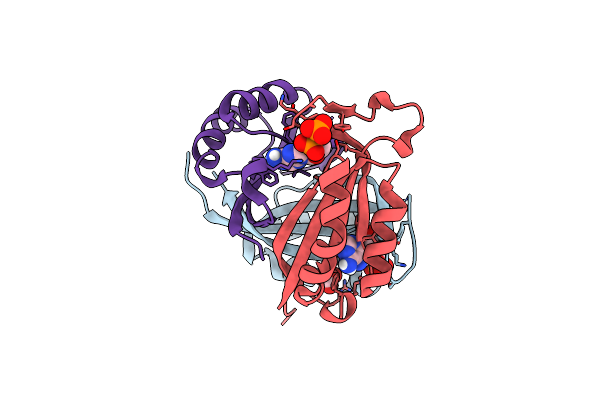
Crystal Structure Of The C-Terminal Catalytic Domain Of Isc1926 Tnpa, An Is607-Like Serine Recombinase
Organism: Sulfolobus sp. l00 11
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2018-07-18 Classification: HYDROLASE |
 |
2.3 Angstrom Structure Of Cpii, A Nitrogen Regulatory Pii-Like Protein From Thiomonas Intermedia K12, Bound To Adp, Amp And Bicarbonate.
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2016-10-12 Classification: METAL BINDING PROTEIN Ligands: AMP, ADP, BCT |
 |
Structure Of The Apo Form Of Cpii From Thiomonas Intermedia K12, A Nitrogen Regulatory Pii-Like Protein
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-09-28 Classification: SIGNALING PROTEIN |
 |
Structure Of Cpii Bound To Adp, Amp And Acetate, From Thiomonas Intermedia K12
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-09-28 Classification: SIGNALING PROTEIN Ligands: ADP, ACT, AMP |
 |
Structure Of Cpii, A Nitrogen Regulatory Pii-Like Protein From Thiomonas Intermedia K12, Bound To Adp, Amp And Bicarbonate.
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-28 Classification: SIGNALING PROTEIN Ligands: ADP, BCT, AMP |
 |
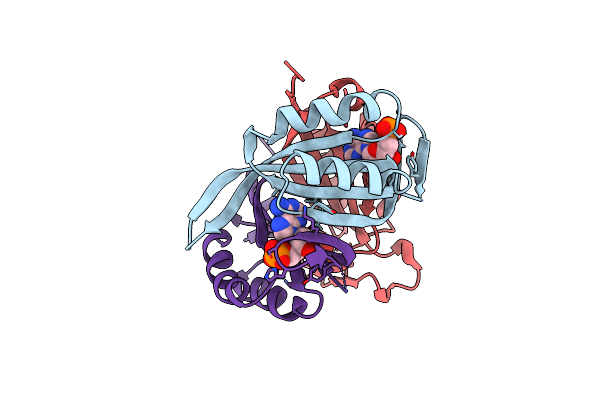
Structure Of Cpii Bound To Adp And Bicarbonate, From Thiomonas Intermedia K12
Organism: Thiomonas arsenitoxydans (strain dsm 22701 / cip 110005 / 3as)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-09-28 Classification: SIGNALING PROTEIN Ligands: ADP, BCT |
 |
2.0 A Structure Of Cpii, A Nitrogen Regulatory Pii-Like Protein From Thiomonas Intermedia K12, Bound Amp
Organism: Thiomonas intermedia (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-09-28 Classification: METAL BINDING PROTEIN Ligands: AMP, CL |
 |
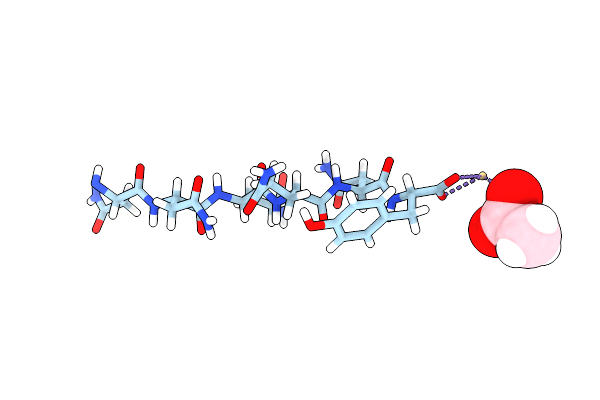
Structure Of Nnqqny From Yeast Prion Sup35 With Zinc Acetate Determined By Microed
Organism: Saccharomyces cerevisiae
Method: ELECTRON CRYSTALLOGRAPHY Resolution:1.00 Å Release Date: 2016-09-14 Classification: PROTEIN FIBRIL Ligands: ZN, ACY |
 |
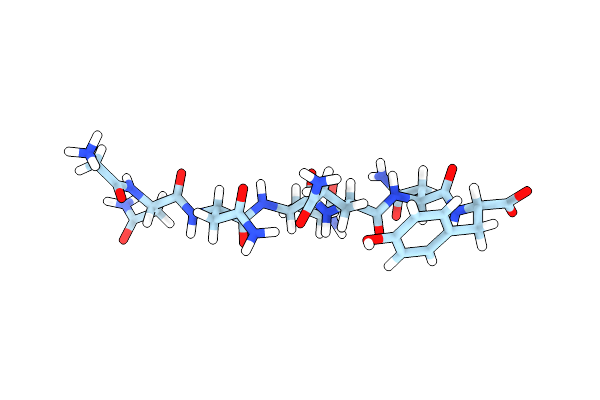
Structure Of Nnqqny From Yeast Prion Sup35 With Cadmium Acetate Determined By Microed
Organism: Saccharomyces cerevisiae
Method: ELECTRON CRYSTALLOGRAPHY Resolution:1.00 Å Release Date: 2016-09-14 Classification: PROTEIN FIBRIL Ligands: CD, ACT |
 |
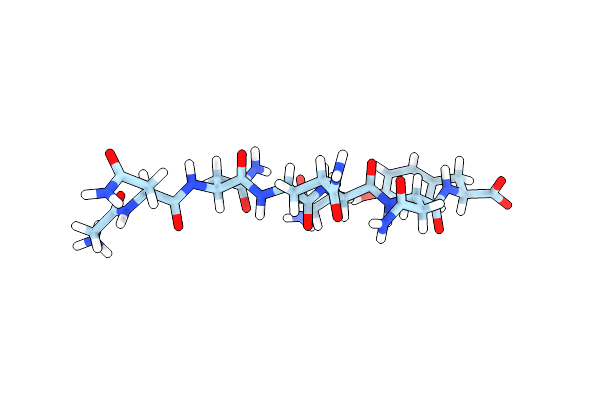
Structure Of Gnnqqny From Yeast Prion Sup35 In Space Group P21 Determined By Microed
Organism: Saccharomyces cerevisiae
Method: ELECTRON CRYSTALLOGRAPHY Resolution:1.10 Å Release Date: 2016-09-14 Classification: PROTEIN FIBRIL |
 |
Structure Of Gnnqqny From Yeast Prion Sup35 In Space Group P212121 Determined By Microed
Organism: Saccharomyces cerevisiae
Method: ELECTRON CRYSTALLOGRAPHY Resolution:1.05 Å Release Date: 2016-09-14 Classification: PROTEIN FIBRIL |
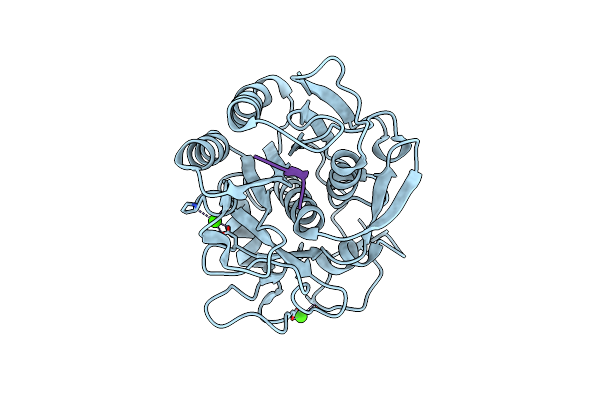 |
Crystal Structure Of Proteinase K From Engyodontium Albuminhibited By Methoxysuccinyl-Ala-Ala-Pro-Phe-Chloromethyl Ketone At 1.15 A Resolution
Organism: Engyodontium album, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2015-05-06 Classification: hydrolase/hydrolase inhibitor Ligands: CA |
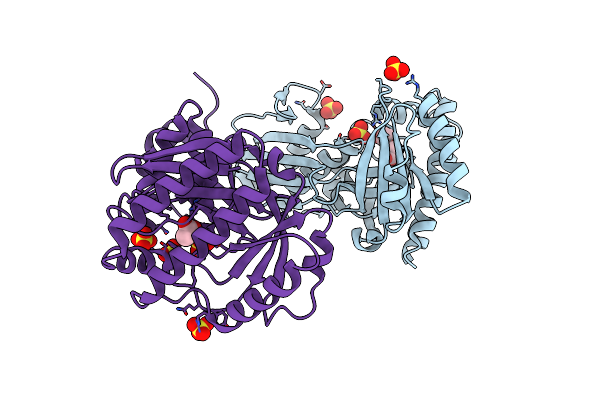 |
Crystal Structure Of Mevalonate-3-Kinase From Thermoplasma Acidophilum (Apo Form)
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: SO4, ACT |
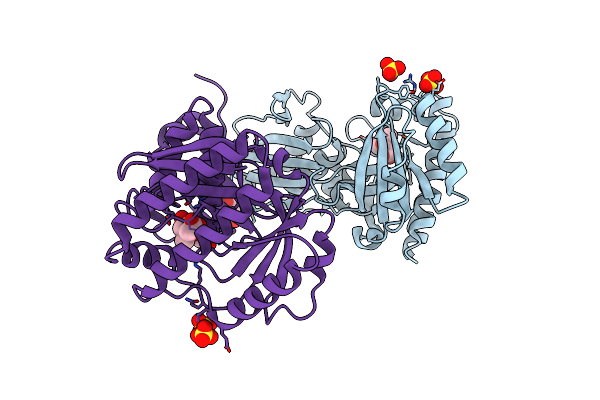 |
Crystal Structure Of Mevalonate-3-Kinase From Thermoplasma Acidophilum (Mevalonate Bound)
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: MEV, SO4, ACT, GOL |

