Search Count: 19
 |
Crystal Structure Of Human Phosphatidylcholine Transfer Protein In Complex With Pc(16:0/20:4)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-04-12 Classification: LIPID BINDING PROTEIN Ligands: M2R, GOL |
 |
Human Start Domain Of Acyl-Coenzyme A Thioesterase 11 (Acot11) Bound To Myristic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2020-09-02 Classification: LIPID BINDING PROTEIN Ligands: MYR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2020-06-03 Classification: APOPTOSIS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2018-12-12 Classification: TRANSFERASE Ligands: G4W |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-01-15 Classification: transferase/transferase inhibitor Ligands: DJK, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.53 Å Release Date: 2014-01-15 Classification: transferase/transferase inhibitor Ligands: YUN |
 |
Crystal Structure Of The Extracellular Domain Of The Epidermal Growth Factor Receptor In Complex With An Adnectin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2012-02-01 Classification: PROTEIN BINDING/SIGNALING PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2012-02-01 Classification: PROTEIN BINDING/CYTOKINE Ligands: HOH |
 |
Crystal And Cryoem Structural Studies Of A Cell Wall Degrading Enzyme In The Bacteriophage Phi29 Tail
Organism: Bacteriophage phi-29
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-07-01 Classification: HYDROLASE Ligands: ZN |
 |
Crystal And Cryoem Structural Studies Of A Cell Wall Degrading Enzyme In The Bacteriophage Phi29 Tail
Organism: Bacteriophage phi-29
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-07-01 Classification: HYDROLASE |
 |
Crystal And Cryoem Structural Studies Of A Cell Wall Degrading Enzyme In The Bacteriophage Phi29 Tail
Organism: Bacteriophage phi-29
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-07-01 Classification: HYDROLASE |
 |
Crystal And Cryoem Structural Studies Of A Cell Wall Degrading Enzyme In The Bacteriophage Phi29 Tail
Organism: Bacteriophage phi-29
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2008-07-01 Classification: HYDROLASE |
 |
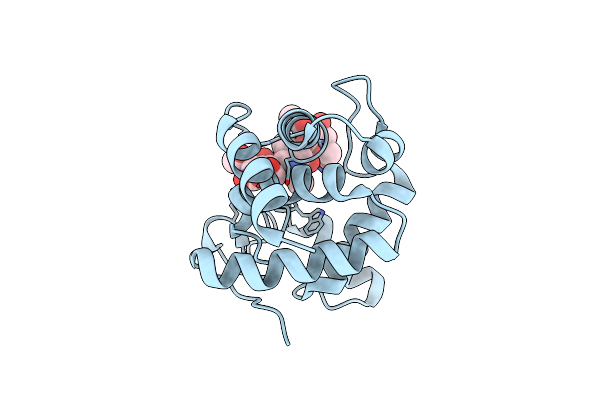
Crystal And Cryoem Structural Studies Of A Cell Wall Degrading Enzyme In The Bacteriophage Phi29 Tail
Organism: Bacteriophage phi-29
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2008-07-01 Classification: HYDROLASE |
 |
Crystal And Cryoem Structural Studies Of A Cell Wall Degrading Enzyme In The Bacteriophage Phi29 Tail
Organism: Bacteriophage phi-29
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2008-07-01 Classification: HYDROLASE |
 |
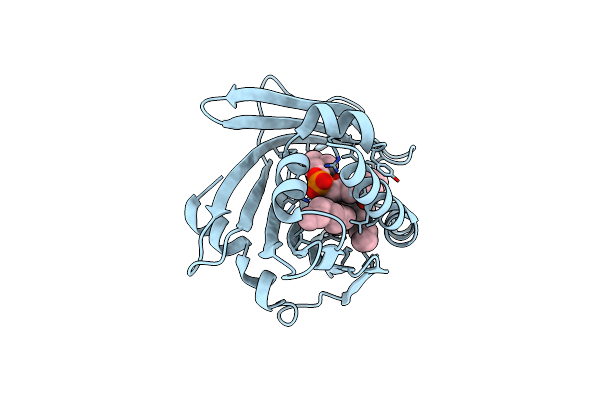
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2004-08-03 Classification: CELL ADHESION |
 |
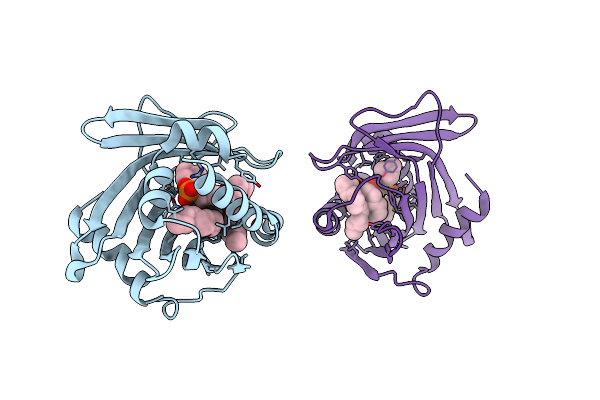
Crystal Structure Of Human Phosphatidylcholine Transfer Protein In Complex With Dilinoleoylphosphatidylcholine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2002-06-26 Classification: LIPID BINDING PROTEIN Ligands: DLP |
 |
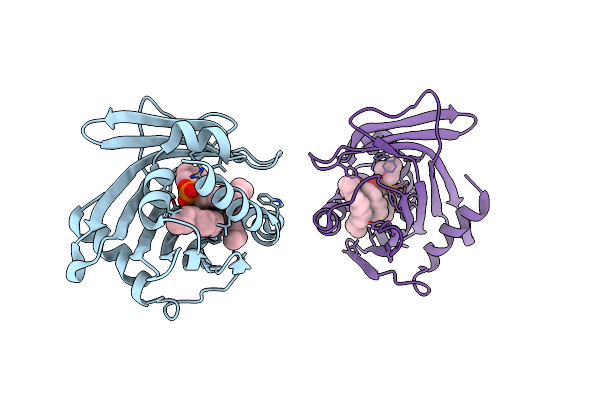
Crystal Structure Of Human Phosphatidylcholine Transfer Protein In Complex With Dilinoleoylphosphatidylcholine (Seleno-Met Protein)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2002-06-26 Classification: LIPID BINDING PROTEIN Ligands: DLP |
 |
Structure Of Human Phosphatidylcholine Transfer Protein In Complex With Palmitoyl-Linoleoyl Phosphatidylcholine (Seleno-Met Protein)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2002-06-26 Classification: LIPID BINDING PROTEIN Ligands: CPL |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2001-03-07 Classification: ISOMERASE Ligands: MN |

