Search Count: 32
 |
Organism: Sinapis alba
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSCRIPTION Ligands: DTT, ZN, FE, CA, K, SAH |
 |
Crystal Structure Of The Plastid Redox Insensitive 2 From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-04-02 Classification: TRANSCRIPTION Ligands: GOL |
 |
Crystal Structure Of The Chloroplastic Fe Superoxide Dismutase Pap9 From Arabidopsis Thaliana.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-07-07 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: PLP, ACT, GOL |
 |
Crystal Structure Of The Prephenate Aminotransferase From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: PLP, NA, CIT |
 |
Crystal Structure Of The Prephenate Aminotransferase From Rhizobium Meliloti
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of The Chloroplastic Gamma-Ketol Reductase From Arabidopsis Thaliana Bound To 13-Oxo-9(Z),11(E),15(Z)- Octadecatrienoic Acid.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE Ligands: KZH |
 |
Crystal Structure Of The Chloroplastic Gamma-Ketol Reductase From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of The Chloroplastic Gamma-Ketol Reductase From Arabidopsis Thaliana Bound To 13Kote And Nadp
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE Ligands: NAP, KZH |
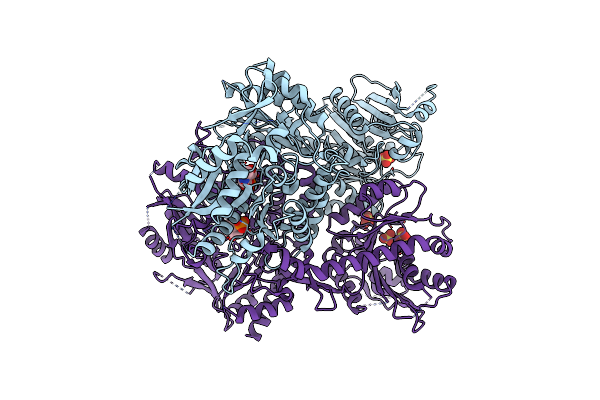 |
Structure Of Selenomethionine Substituted Bifunctional Dapa Aminotransferase-Dethiobiotin Synthetase From Arabidopsis Thaliana In Its Apo Form.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2012-06-13 Classification: TRANSFERASE Ligands: PLP, SO4 |
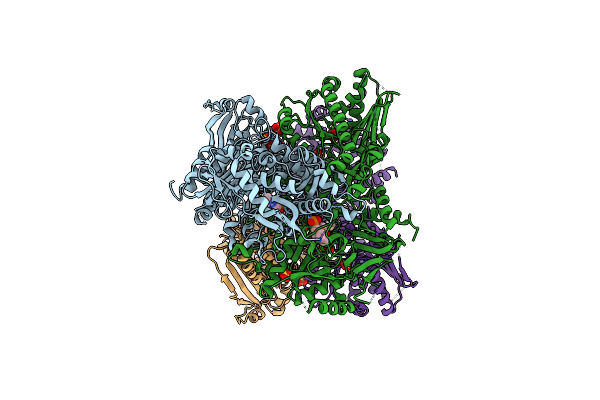 |
Structure Of Bifunctional Dapa Aminotransferase-Dtb Synthetase From Arabidopsis Thaliana In Its Apo Form.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-06-13 Classification: TRANSFERASE Ligands: PLP, SO4, MG |
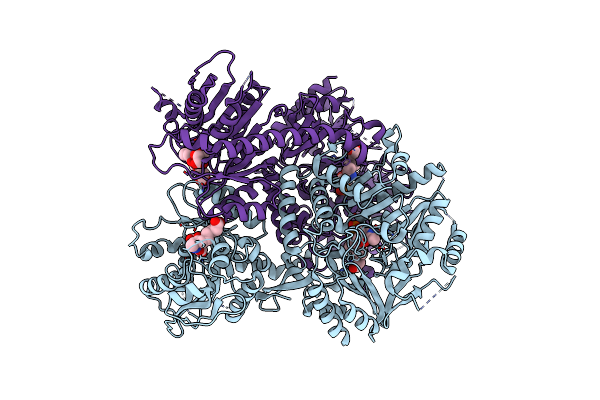 |
Structure Of Bifunctional Dapa Aminotransferase-Dtb Synthetase From Arabidopsis Thaliana Bound To 7-Keto 8-Amino Pelargonic Acid (Kapa)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2012-06-13 Classification: TRANSFERASE Ligands: PLP, KAP, TLA |
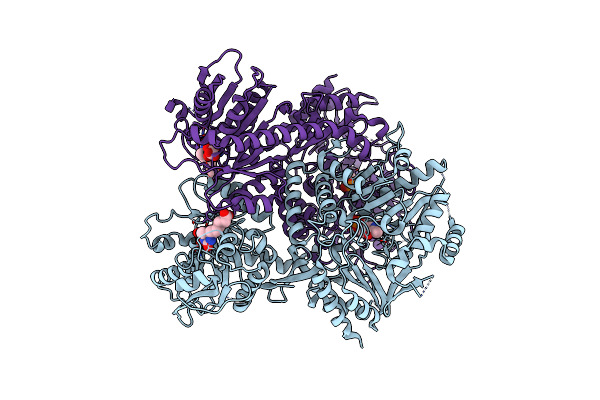 |
Structure Of Bifunctional Dapa Aminotransferase-Dtb Synthetase From Arabidopsis Thaliana Bound To Dethiobiotin (Dtb).
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2012-06-13 Classification: TRANSFERASE Ligands: PLP, TLA, DTB |
 |
Enantiopyochelin Outer Membrane Tonb-Dependent Transporter From Pseudomonas Fluorescens Bound To The Ferri-Enantiopyochelin
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:3.26 Å Release Date: 2011-12-07 Classification: METAL TRANSPORT Ligands: SO4, CIT, EFE |
 |
Organism: Synechocystis
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2010-06-09 Classification: TRANSFERASE Ligands: THR, LYS, SO4 |
 |
Crystal Structure Of The Heme/Hemoglobin Outer Membrane Transporter Shua From Shigella Dysenteriae
Organism: Shigella dysenteriae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2009-07-14 Classification: MEMBRANE PROTEIN Ligands: BOG, PB |
 |
Structure Of The Alcaligin Outer Membrane Recepteur Faua From Bordetella Pertussis
Organism: Bordetella pertussis
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2009-03-31 Classification: MEMBRANE PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The Full Length Ferric Pyoverdine Outer Membrane Receptor Fpva Of Pseudomonas Aeruginosa In Its Apo Form
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2007-10-30 Classification: TRANSPORT PROTEIN Ligands: PO4, N8E |
 |
Crystal Structure Of The Ferripyoverdine Receptor Of The Outer Membrane Of Pseudomonas Aeruginosa Bound To Ferripyoverdine.
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2007-09-11 Classification: MEMBRANE PROTEIN Ligands: SO4, PVE, FE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-12-20 Classification: OXIDOREDUCTASE Ligands: FAD, TEO, UNL, FES, SF4, F3S, UQ, GOL, JZR, HEM, PEE |

