Search Count: 251
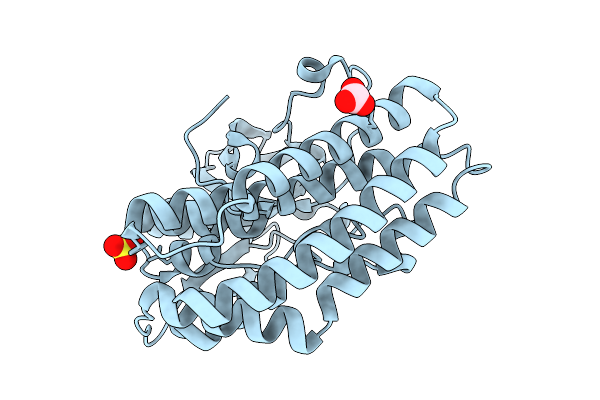 |
Crystal Structure Of Ctag From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
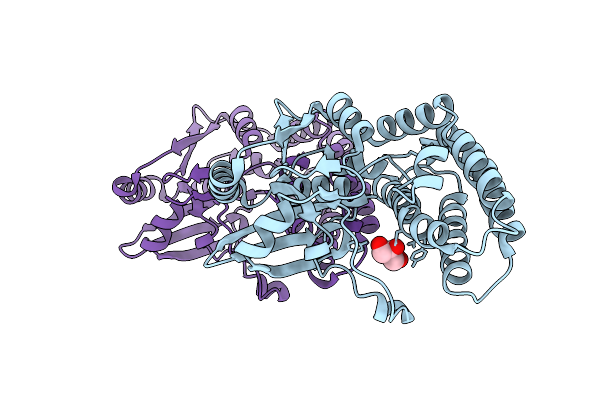 |
Crystal Structure Of Ctag From Ruminiclostridium Cellulolyticum (P2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: GOL |
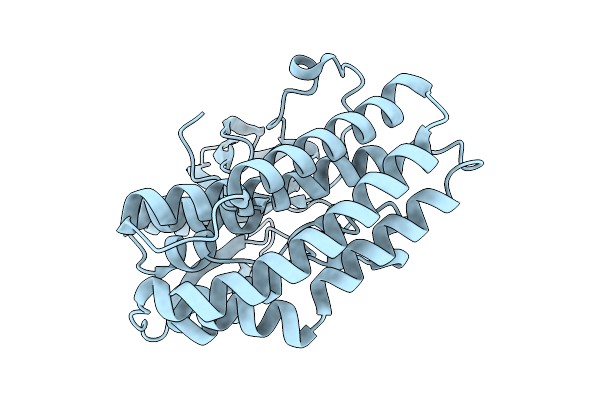 |
Crystal Structure Of The Ctag_C11A Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-10-01 Classification: LIGASE |
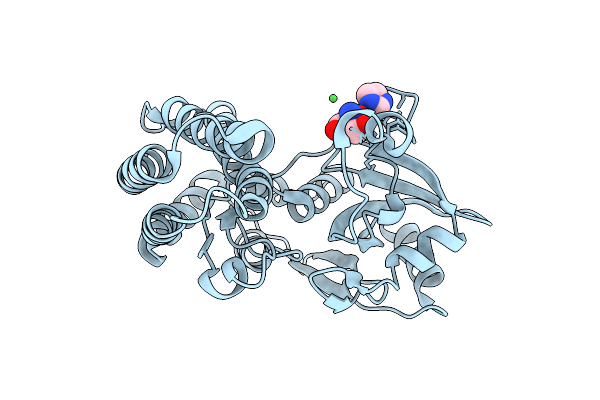 |
Crystal Structure Of The Ctag_C11A Variant From Ruminiclostridium Cellulolyticum (P2(1)2(1)2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: NI, IMD, TRS |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum (P2(1)2(1)2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-10-01 Classification: LIGASE |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum In Complex With Phba (P2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: HBA, PO4 |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum In Complex With Phba (P2(1)2(1)2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: HBA, GOL |
 |
Crystal Structure Of The Ctag_D144N Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-10-01 Classification: LIGASE |
 |
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-11-09 Classification: LIPID BINDING PROTEIN Ligands: IQ4, ZN, NA, TRS |
 |
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-11-09 Classification: LIPID BINDING PROTEIN Ligands: GOL, NA |
 |
The Crystal Structure Of Clostridium Cellulolyticum Laci Family Transcriptional Regulator Ccel_1438
Organism: Ruminiclostridium cellulolyticum h10
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-07-27 Classification: TRANSCRIPTION |
 |
Organism: Ruminiclostridium cellulolyticum h10
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2022-07-27 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Organism: Ruminiclostridium cellulolyticum (strain atcc 35319 / dsm 5812 / jcm 6584 / h10)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-03-24 Classification: ISOMERASE Ligands: MN, GOL |
 |
Gh5-4 Broad Specificity Endoglucanase From Hungateiclostridium Cellulolyticum
Organism: Hungateiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-11-13 Classification: HYDROLASE Ligands: PG4, PEG, PGE |
 |
Crystal Structures Of D-Psicose 3-Epimerase From Clostridium Cellulolyticum H10 And Its Complex With Ketohexose Sugars
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2012-08-01 Classification: ISOMERASE Ligands: MN |
 |
Crystal Structures Of D-Psicose 3-Epimerase With D-Psicose From Clostridium Cellulolyticum H10
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2012-08-01 Classification: ISOMERASE Ligands: PSJ, MN |
 |
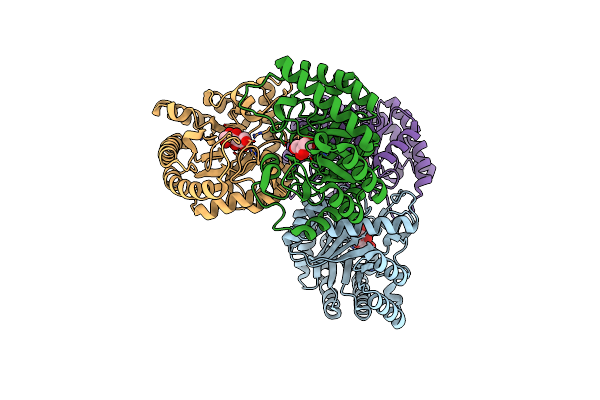
Crystal Structures Of D-Psicose 3-Epimerase With D-Fructose From Clostridium Cellulolyticum H10
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2012-08-01 Classification: ISOMERASE Ligands: FUD, MN |
 |
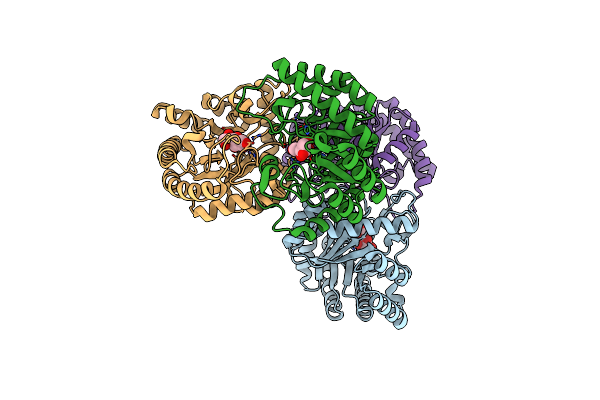
Crystal Structures Of D-Psicose 3-Epimerase With D-Tagatose From Clostridium Cellulolyticum H10
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-08-01 Classification: ISOMERASE Ligands: TAG, MN |
 |
Crystal Structures Of D-Psicose 3-Epimerase With D-Sorbose From Clostridium Cellulolyticum H10
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2012-08-01 Classification: ISOMERASE Ligands: SDD, MN |

