Search Count: 373
 |
Organism: Clostridium beijerinckii
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: ZN, SF4, 402 |
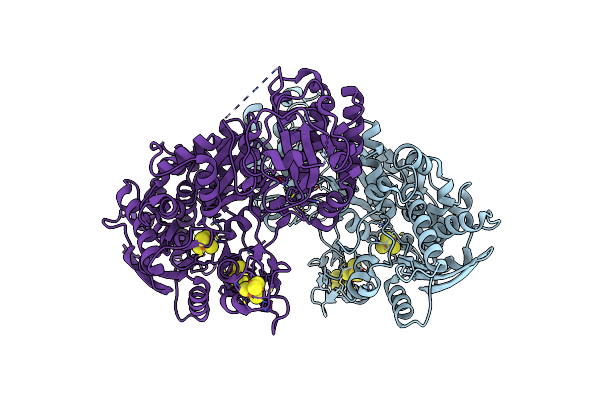 |
Crystal Structure Of Apo-[Fefe]-Hydrogenase Cba5H From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: SF4, ZN, CL |
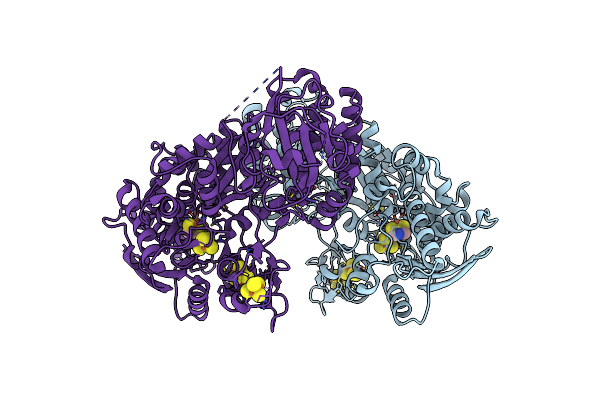 |
Crystal Structure Of [Fefe]-Hydrogenase Cba5H From Clostridium Beijerinckii In Hinact State
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: 402, SF4, ZN, CL |
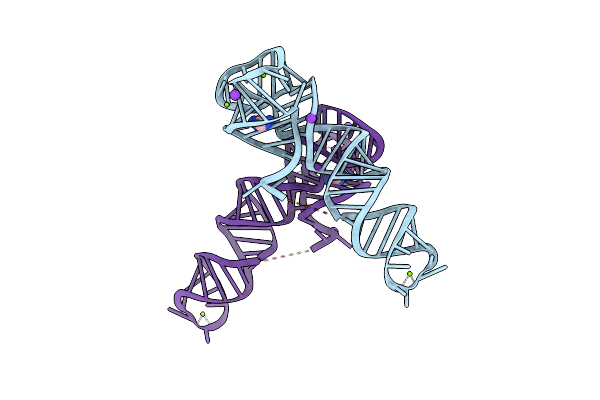 |
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-11-27 Classification: RNA Ligands: MG, 5AC, K |
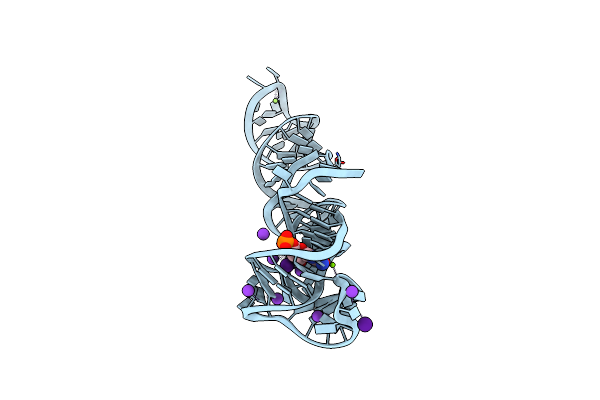 |
Cocrystal Structure Of Clostridium Beijerinckii Ztp Riboswitch With Zmp And Cs
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2024-11-27 Classification: RNA Ligands: CS, MG, AMZ, K |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase Gapdh From Clostridium Beijerinckii
Organism: Clostridium beijerinckii ncimb 8052
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-03-17 Classification: OXIDOREDUCTASE Ligands: BME, MG |
 |
Crystal Structure Of [Fefe]-Hydrogenase Cba5H (Partial) From Clostridium Beijerinckii In Hinact State
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-12-09 Classification: OXIDOREDUCTASE Ligands: 402, SF4 |
 |
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-26 Classification: OXIDOREDUCTASE Ligands: NAD, 2PE, ZN, MG |
 |
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-05-29 Classification: OXIDOREDUCTASE Ligands: NDP, EDO |
 |
Crystal Structure Of A Beta-1,3-Glucanase Domain (Gh64) From Clostridium Beijerinckii
Organism: Clostridium beijerinckii ncimb 8052
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-11-08 Classification: HYDROLASE |
 |
Organism: Clostridium beijerinckii (strain atcc 51743 / ncimb 8052)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-08-02 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Clostridium beijerinckii (strain atcc 51743 / ncimb 8052)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-02 Classification: SUGAR BINDING PROTEIN Ligands: XYP |
 |
Organism: Clostridium beijerinckii (strain atcc 51743 / ncimb 8052)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-08-02 Classification: SUGAR BINDING PROTEIN Ligands: XYP |
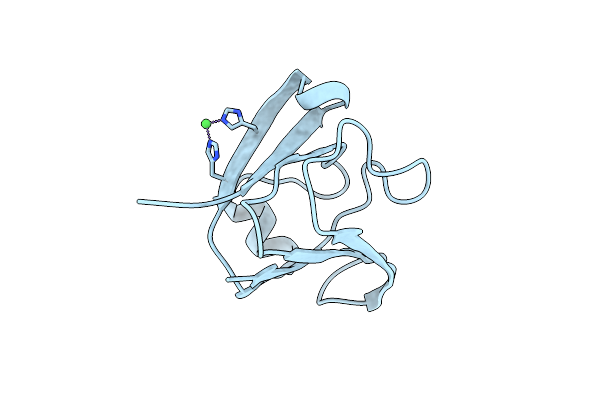 |
Crystal Structure Of Clostrillin Double Mutant (S17H,S19H) In Complex With Nickel
Organism: Clostridium beijerinckii ncimb 8052
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-02-01 Classification: METAL BINDING PROTEIN Ligands: NI |
 |
Crystal Structure Of Enolase Superfamily Member From Clostridium Beijerincki Complexed With Mg
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-05-23 Classification: ISOMERASE Ligands: MG |
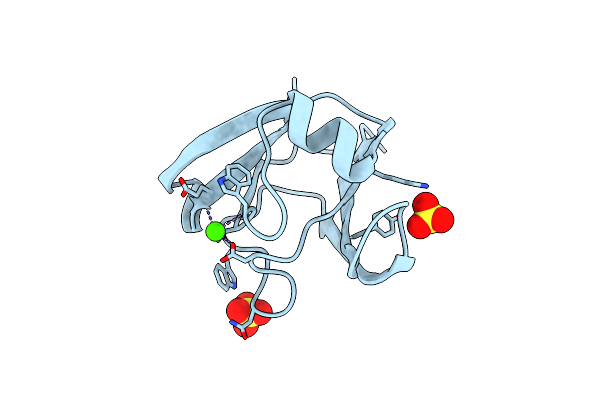 |
Crystal Structure Of A Mutant T82R Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-11-16 Classification: METAL BINDING PROTEIN Ligands: CA, SO4 |
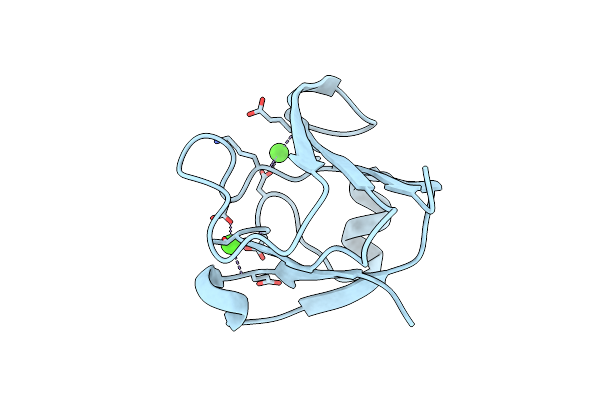 |
Crystal Structure Of A Mutant W39D Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-11-16 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of A Mutant T41S Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2011-11-16 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of A Double Mutant T41S T82S Of A Betagamma-Crystallin Domain From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-11-16 Classification: METAL BINDING PROTEIN Ligands: CA, SO4 |
 |
Q165E/S254K Double Mutant Chimera Of Alcohol Dehydrogenase By Exchange Of The Cofactor Binding Domain Res 153-295 Of T. Brockii Adh By C. Beijerinckii Adh
Organism: Thermoanaerobacter brockii, Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2010-01-26 Classification: OXIDOREDUCTASE Ligands: ZN, ACT, EDO, CL |

