Search Count: 550
All
Selected
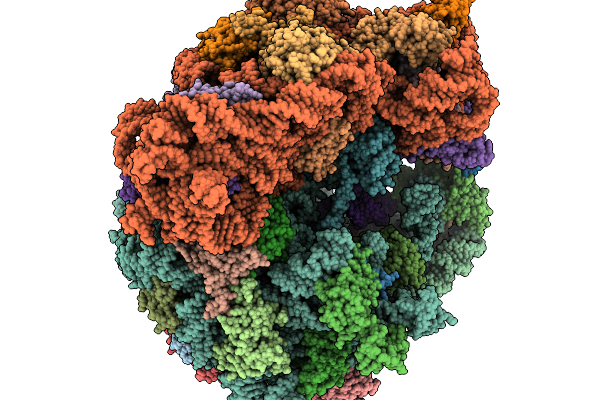 |
Structure Of Clim-Stalled Bacillus Subtilis 70S Ribosome With Release Factor Bound In The A-Site
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:2.30 Å Release Date: 2026-01-14 Classification: RIBOSOME Ligands: ZN |
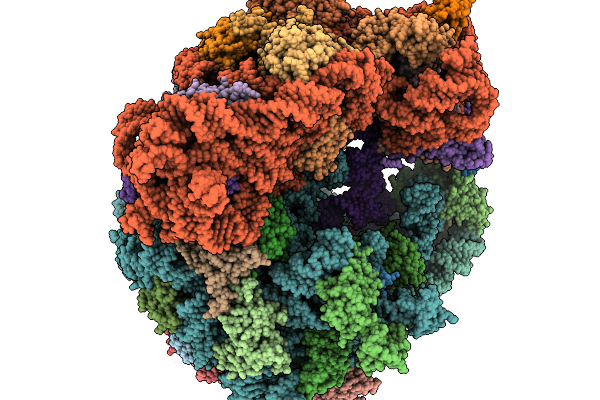 |
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:2.30 Å Release Date: 2026-01-14 Classification: RIBOSOME |
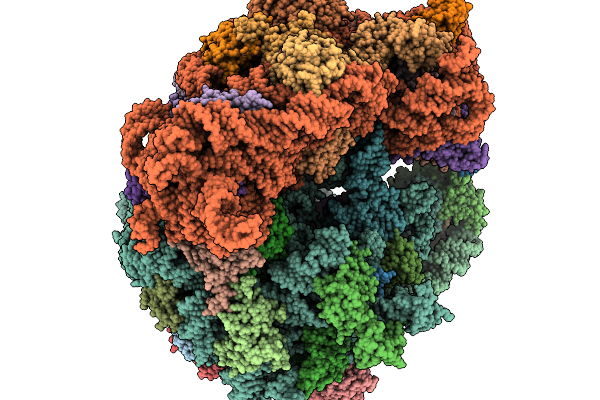 |
Structure Of Clim-Stalled Bacillus Subtilis 70S Ribosome With Trna-Tyr In The A-Site
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2026-01-14 Classification: RIBOSOME Ligands: ZN |
 |
Cryoem Structure Of De Novo Antibody Fragment Scfv 6 With C. Difficile Toxin B (Tcdb)
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN |
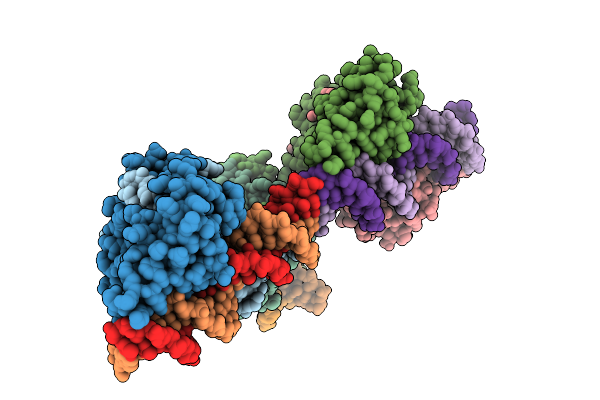 |
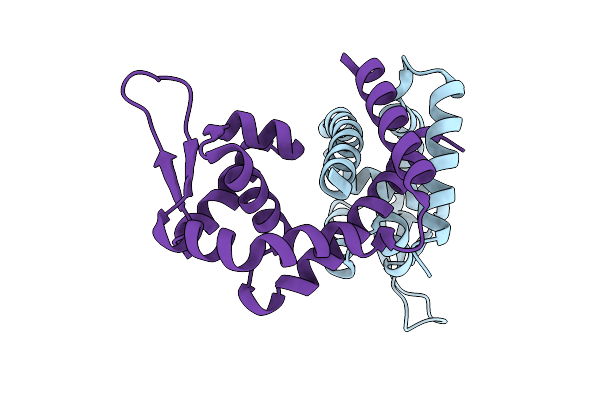
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: K |
 |
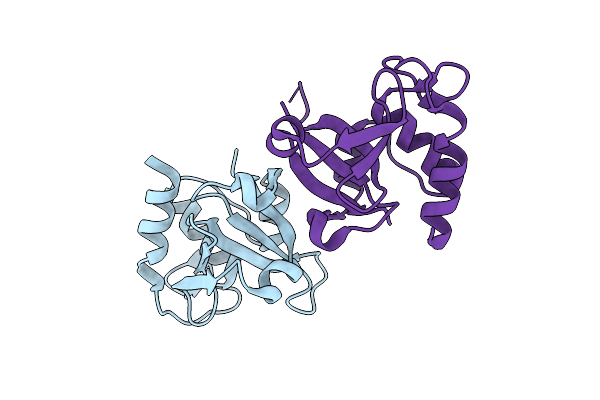
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-10-29 Classification: TRANSCRIPTION |
 |
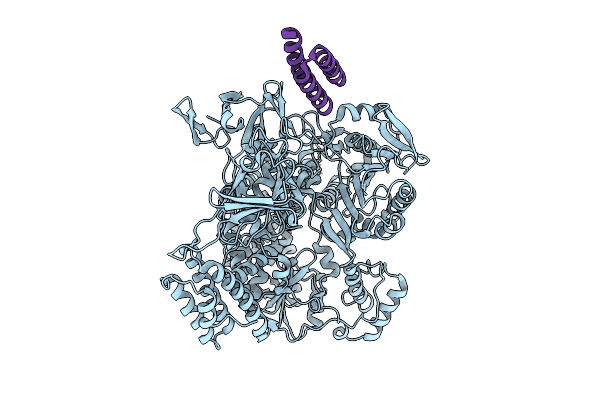
X-Ray Structure Of Clostridioides Difficile Autolysin Acd33800 Catalytic Domain
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-10-15 Classification: ANTIMICROBIAL PROTEIN |
 |
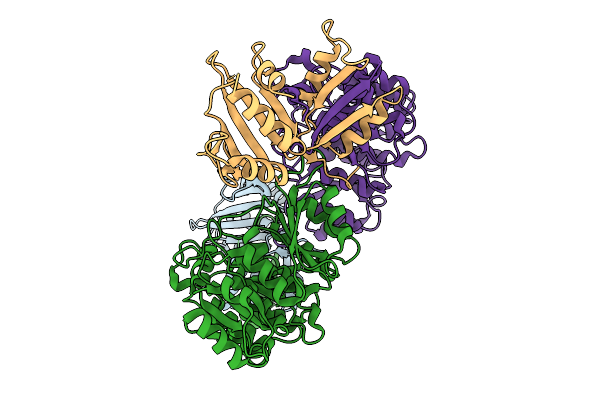
De Novo Designed Minibinder Complexed With Clostridioides Difficile Toxin B
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: TOXIN |
 |
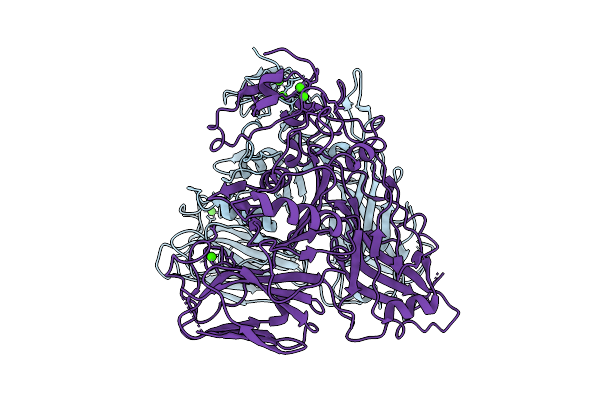
Slpl/Slph (H/L) Complex From C. Difficile Slpa (Ox247_Delta_Orf2 Strain, Slct11)
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-09-03 Classification: STRUCTURAL PROTEIN |
 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2025-09-03 Classification: STRUCTURAL PROTEIN |
 |
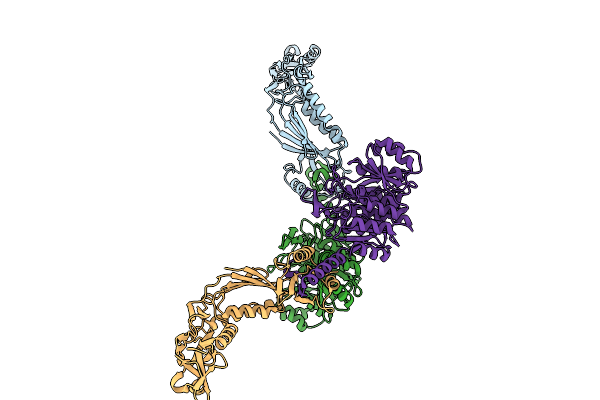
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-08-20 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridioides difficile r20291
Method: ELECTRON MICROSCOPY Resolution:3.56 Å Release Date: 2025-07-16 Classification: TOXIN Ligands: CA |
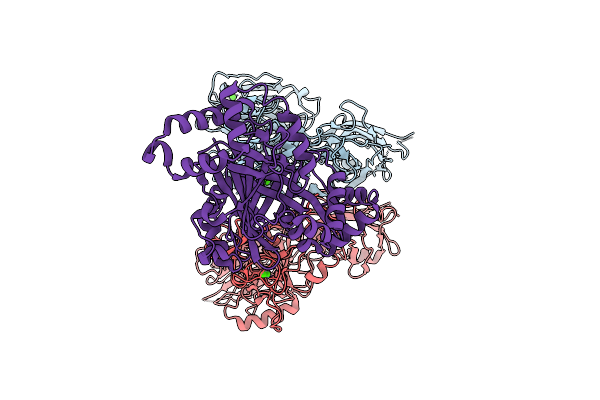 |
Clostridioides Difficile Transferase B Component Dimer In Complex With The A Component
Organism: Clostridioides difficile r20291
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TOXIN Ligands: CA |
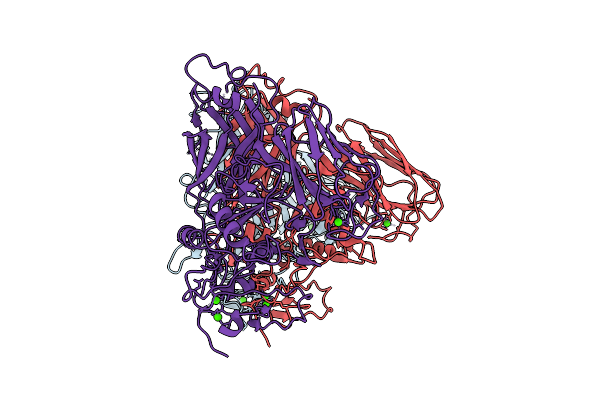 |
Organism: Clostridioides difficile r20291
Method: ELECTRON MICROSCOPY Resolution:4.19 Å Release Date: 2025-07-16 Classification: TOXIN Ligands: CA |
 |
Clostridioides Difficile Transferase B Component Trimer In Complex With The A Component
Organism: Clostridioides difficile r20291
Method: ELECTRON MICROSCOPY Resolution:7.86 Å Release Date: 2025-07-16 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridioides difficile r20291
Method: ELECTRON MICROSCOPY Resolution:7.01 Å Release Date: 2025-07-16 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridioides difficile r20291
Method: ELECTRON MICROSCOPY Resolution:3.33 Å Release Date: 2025-07-16 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridioides difficile, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-07-09 Classification: TRANSFERASE,TOXIN/IMMUNE SYSTEM |
 |
Crystal Structure Of The Surface Protein (Cd630_07380) From Clostridium Difficile Strain 630
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN |
 |
Organism: Clostridioides difficile p28
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: FE, O |

