Search Count: 299
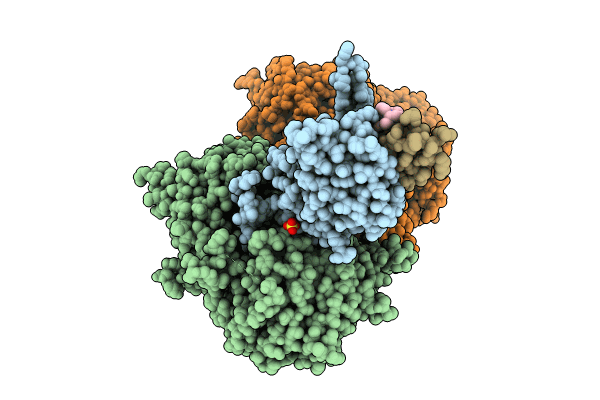 |
Ternary Complex Structure Of Cereblon-Ddb1 Bound To Wiz(Zf7) And The Molecular Glue Dwiz-1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-07-10 Classification: LIGASE Ligands: ZN, U3I, SO4, EDO |
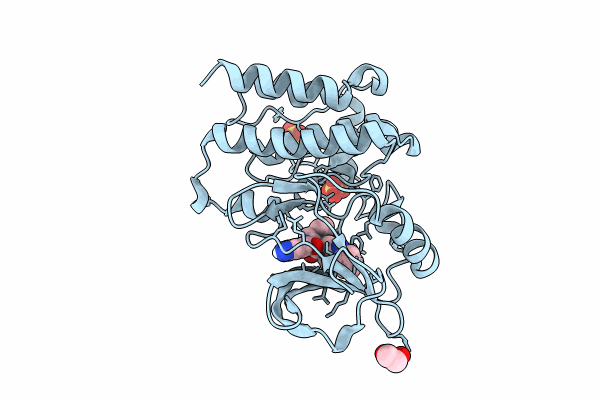 |
Bruton'S Tyrosine Kinase In Complex With N-[(2R)-1-[(3R)-3-(Methylcarbamoyl)-1H,2H,3H,4H,9H-Pyrido[3,4-B]Indol-2-Yl]-3-(3-Methylphenyl)-1-Oxopropan-2-Yl]-1H-Indazole-5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: UQX, NA, CL, SO4, EDO |
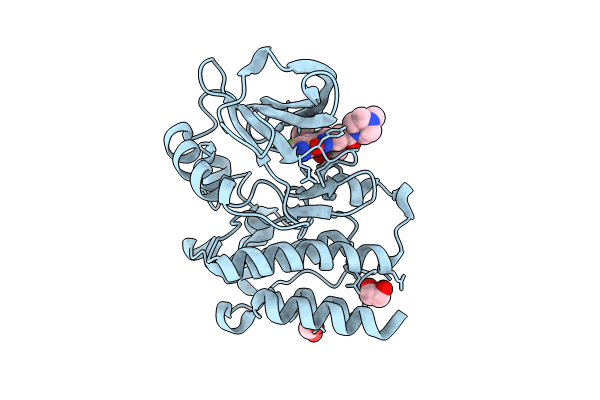 |
Bruton'S Tyrosine Kinase In Complex With N-[(2R)-1-[(3R)-3-(Methylcarbamoyl)-1H,2H,3H,4H,9H-Pyrido[3,4-B]Indol-2-Yl]-3-(3-Methylphenyl)-1-Oxopropan-2-Yl]-1H-Indazole-5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: UP9, EDO, CL, IOD |
 |
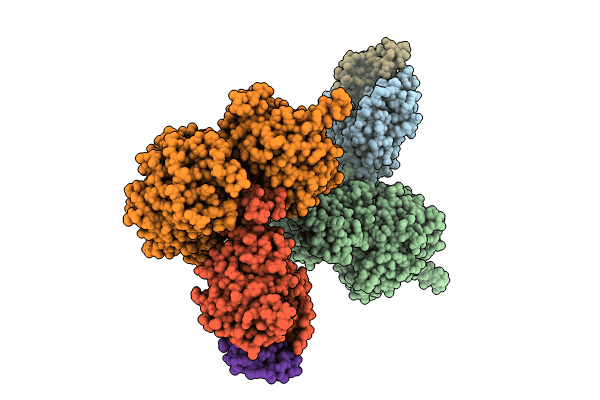
The Ternary Complex Structure Of Ddb1-Crbn-Sall4(Zf1,2)-Short Bound To Cc-220
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2023-12-27 Classification: LIGASE Ligands: ZN, 8W7, SO4 |
 |
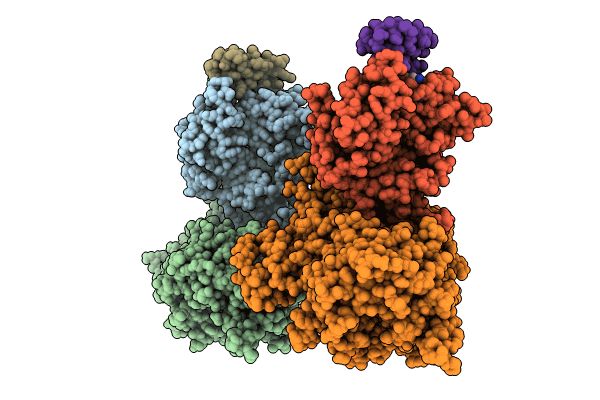
The Ternary Complex Structure Of Ddb1-Crbn-Sall4(Zf1,2)-Short Bound To Pomalidomide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-12-27 Classification: LIGASE Ligands: ZN, Y70, EDO |
 |
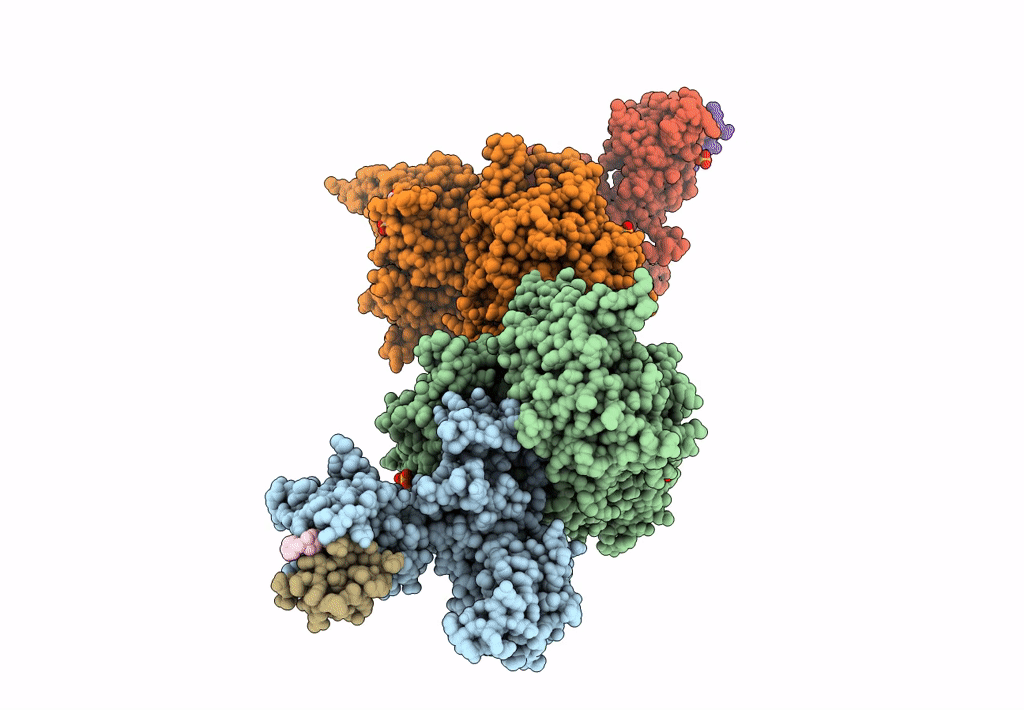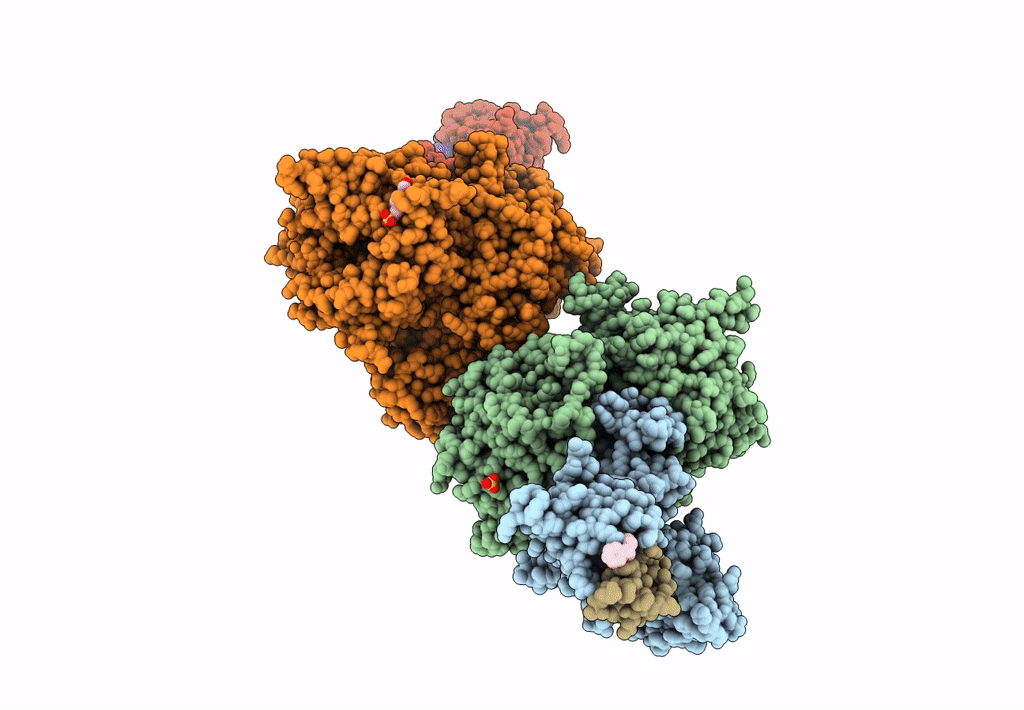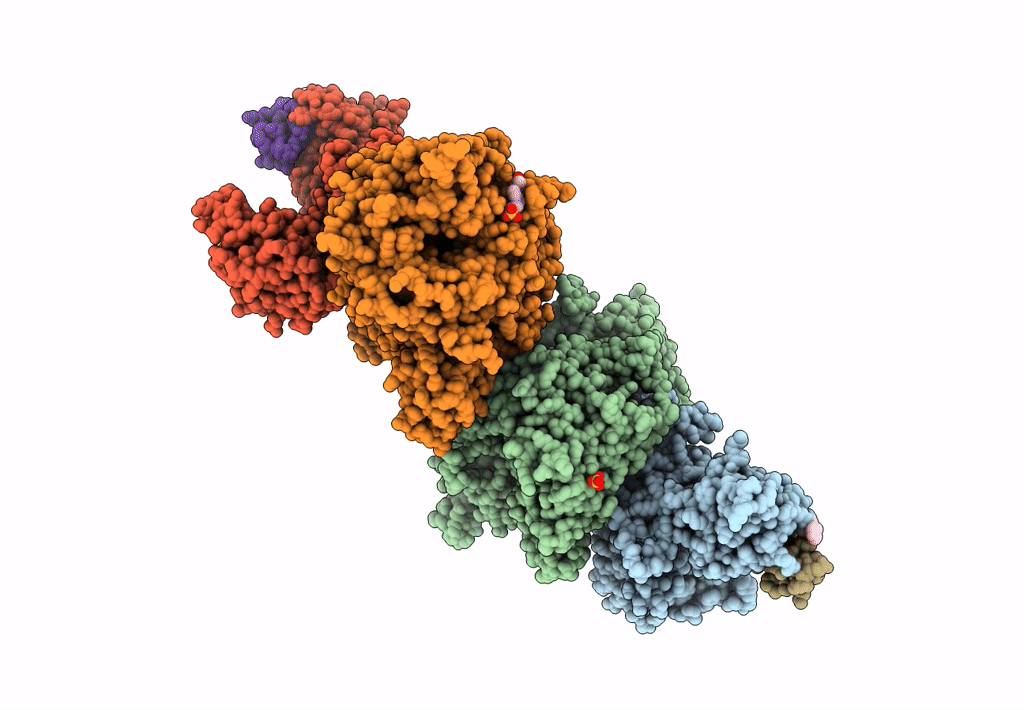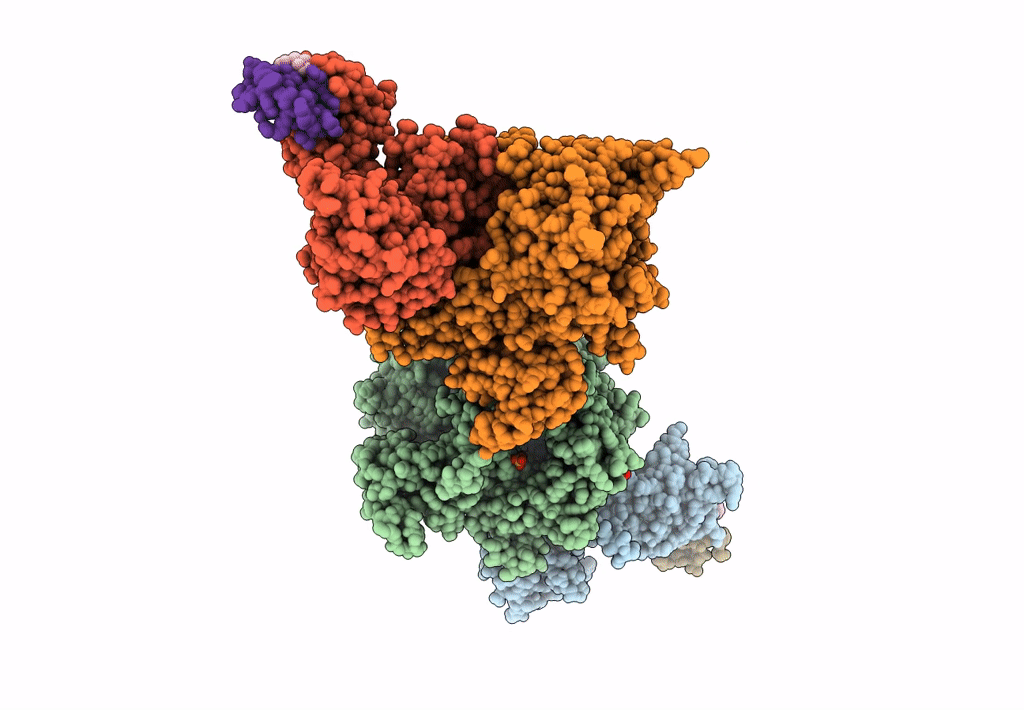
The Ternary Complex Structure Of Ddb1-Crbn-Sall4(Zf1,2)-Long Bound To Pomalidomide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-12-27 Classification: LIGASE Ligands: ZN, Y70 |
 |
Ternary Complex Structure Of Cereblon-Ddb1 Bound To Ikzf2(Zf2) And The Molecular Glue Dky709
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2023-03-08 Classification: LIGASE Ligands: ZN, LWK, SO4, EPE |
 |
Ternary Complex Structure Of Cereblon-Ddb1 Bound To Ikzf2(Zf2,3) And The Molecular Glue Dky709
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2023-03-08 Classification: LIGASE Ligands: ZN, LWK |
 |
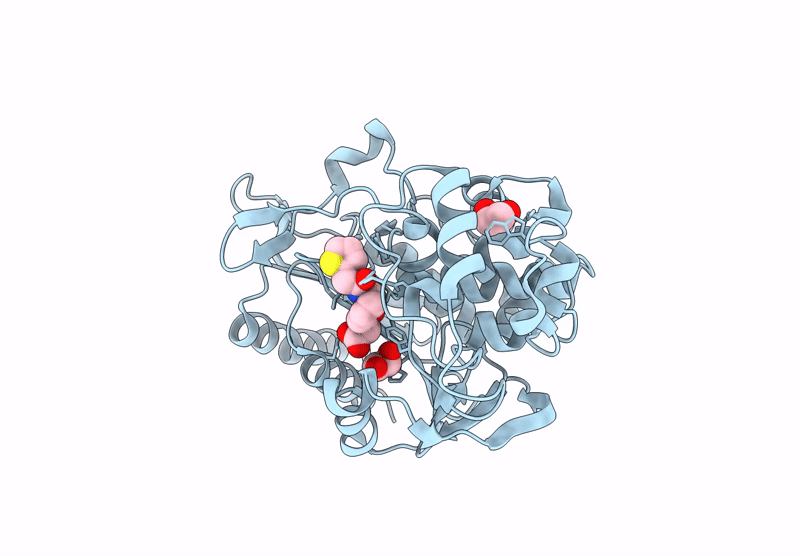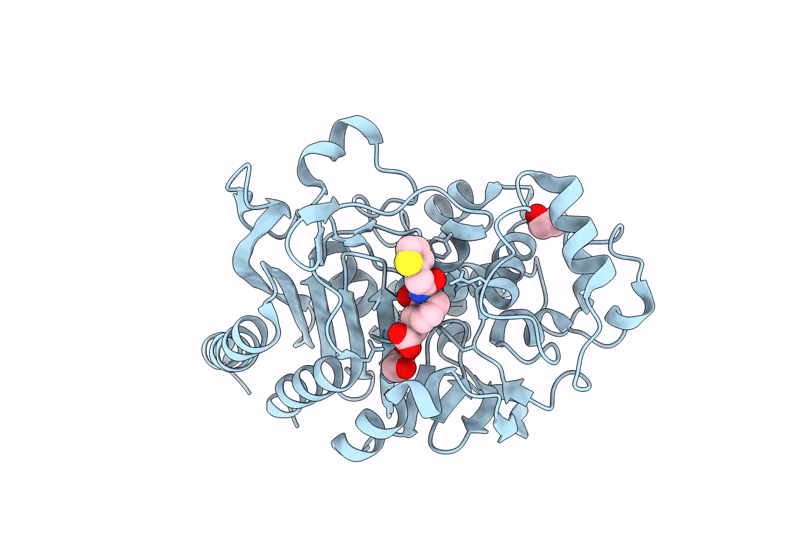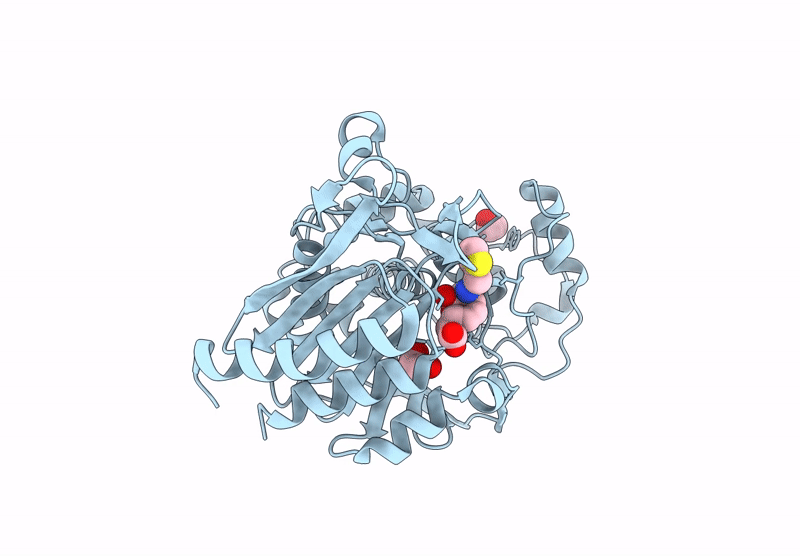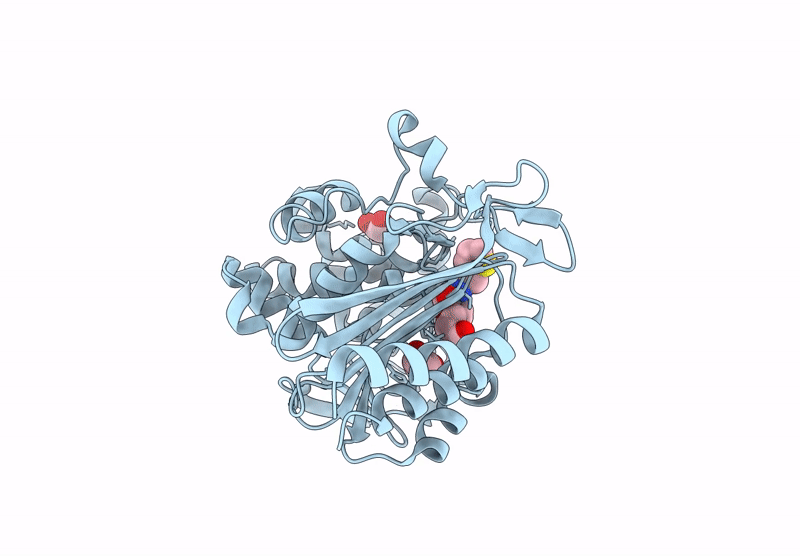
Bruton'S Tyrosine Kinase In Complex With 3-{[4-(1-Acetylpiperidin-4-Yl)Phenyl]Amino}-5-[(3R)-3-(3-Methyl-2-Oxoimidazolidin-1-Yl)Piperidin-1-Yl]Pyrazine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: WKC, EDO, PD, CL |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: ZXQ, EDO, CL |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-10-12 Classification: HYDROLASE Ligands: ZXQ, EDO |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-05-18 Classification: HYDROLASE Ligands: ZXQ, SO4, EDO, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-09-11 Classification: TRANSPORT PROTEIN Ligands: SO4, GOL |
 |
Crystal Structure Of A 3-Oxoacyl-Acyl-Carrier Protein Reductase Fabg4 From Mycobacterium Smegmatis Bound To Nad
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-05-17 Classification: OXIDOREDUCTASE Ligands: NAD, CL |
 |
Crystal Structure Of Phosphinothricin N-Acetyltransferase From Brucella Ovis
Organism: Brucella ovis (strain atcc 25840 / 63/290 / nctc 10512)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-12-02 Classification: TRANSFERASE Ligands: EDO, IMD, GOL |
 |
Organism: Zaire ebolavirus (strain mayinga-76)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-11-04 Classification: VIRAL PROTEIN Ligands: EDO |
 |
Crystal Structure Of Phosphinothricin N-Acetyltransferase From Brucella Ovis In Complex With Acetylcoa <Structure_Details =
Organism: Brucella ovis (strain atcc 25840 / 63/290 / nctc 10512)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-10-28 Classification: TRANSFERASE Ligands: ACO, CL, MG |
 |
Structure Of An Adp Ribosylation Factor From Entamoeba Histolytica Hm-1:Imss Bound To Mg-Gdp
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-05-20 Classification: SIGNALING PROTEIN Ligands: GDP, MG |
 |
Structure Of Udp-N-Acetylmuramoylalanyl-D-Glutamyl-2,6-Diaminopimelate--D-Alanyl-D-Alanyl Ligase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii ab5075
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-05-20 Classification: LIGASE Ligands: ANP, MG, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-05-06 Classification: Apoptosis/Inhibitor Ligands: ZN, 865, POP |

