Search Count: 15
 |
Organism: Streptomyces griseofuscus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-12-28 Classification: TRANSFERASE Ligands: PO4, K, FMT |
 |
C-Methyltransferase Psmd From Streptomyces Griseofuscus With Bound Cofactor (Crystal Form 2)
Organism: Streptomyces griseofuscus
Method: X-RAY DIFFRACTION Release Date: 2022-12-28 Classification: TRANSFERASE Ligands: PO4, MPD, SAH |
 |
C-Methyltransferase Psmd From Streptomyces Griseofuscus With Bound Cofactor (Crystal Form 1)
Organism: Streptomyces griseofuscus
Method: X-RAY DIFFRACTION Release Date: 2022-12-28 Classification: TRANSFERASE Ligands: UNL, PGE, SAH |
 |
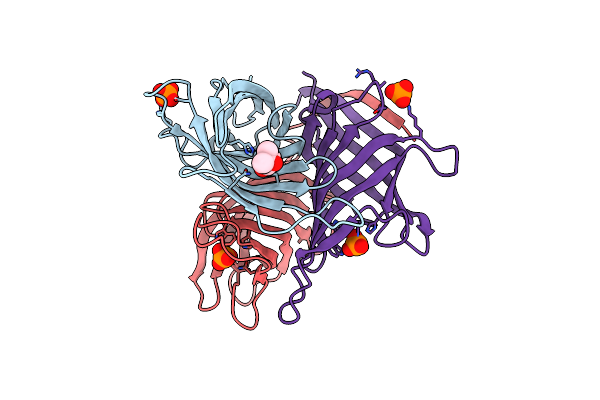
Monocot Chimeric Jacalin Jac1 From Oryza Sativa: Dirigent Domain (Crystal Form 1)
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-05-04 Classification: PLANT PROTEIN Ligands: CA, CL |
 |
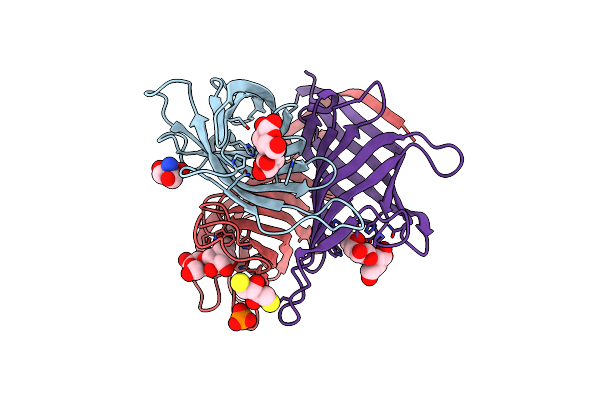
Monocot Chimeric Jacalin Jac1 From Oryza Sativa: Dirigent Domain (Crystal Form 2)
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-05-04 Classification: PLANT PROTEIN Ligands: PO4, MPD |
 |
Monocot Chimeric Jacalin Jac1 From Oryza Sativa: Dirigent Domain With Bound Galactobiose
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-05-04 Classification: PLANT PROTEIN Ligands: TRS, PO4, DTU |
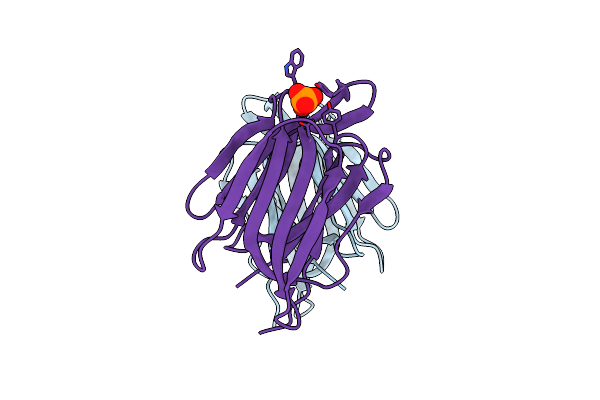 |
Monocot Chimeric Jacalin Jac1 From Oryza Sativa: Lectin Domain (Crystal Form 1)
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-05-04 Classification: PLANT PROTEIN Ligands: PO4 |
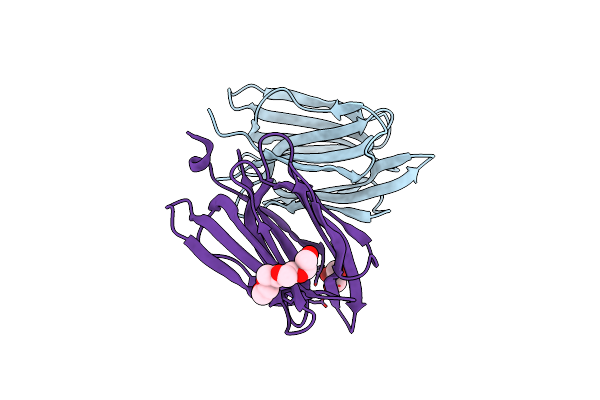 |
Monocot Chimeric Jacalin Jac1 From Oryza Sativa: Lectin Domain (Crystal Form 2)
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-05-04 Classification: PLANT PROTEIN Ligands: IOD, 1PE, GOL |
 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase From Escherichia Coli (K58E-Y96W Mutant)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2016-05-04 Classification: LYASE Ligands: BU2, EPE |
 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase From Escherichia Coli (K58E-Y96W Mutant) After Acetaldehyde Treatment
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2016-05-04 Classification: LYASE Ligands: 1BO, EPE |
 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase From Escherichia Coli (K58E-Y96W Mutant) After Acetaldehyde Treatment And Heating
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-05-04 Classification: LYASE Ligands: 1BO, EPE |
 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase From Colwellia Psychrerythraea (Tetragonal Form)
Organism: Colwellia psychrerythraea
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2016-02-03 Classification: LYASE Ligands: UNL, SO4, CO3 |
 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase From Colwellia Psychrerythraea (Hexagonal Form)
Organism: Colwellia psychrerythraea (strain 34h / atcc baa-681)
Method: X-RAY DIFFRACTION Release Date: 2016-02-03 Classification: LYASE Ligands: UNL, GOL |
 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase From Shewanella Halifaxensis
Organism: Shewanella halifaxensis
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-02-03 Classification: LYASE |
 |
Crystal Structure Of 2-Keto-3-Deoxy-6-Phospho-Gluconate Aldolase From Zymomonas Mobilis Atcc 29191
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2014-04-30 Classification: LYASE Ligands: SO4 |

