Search Count: 4
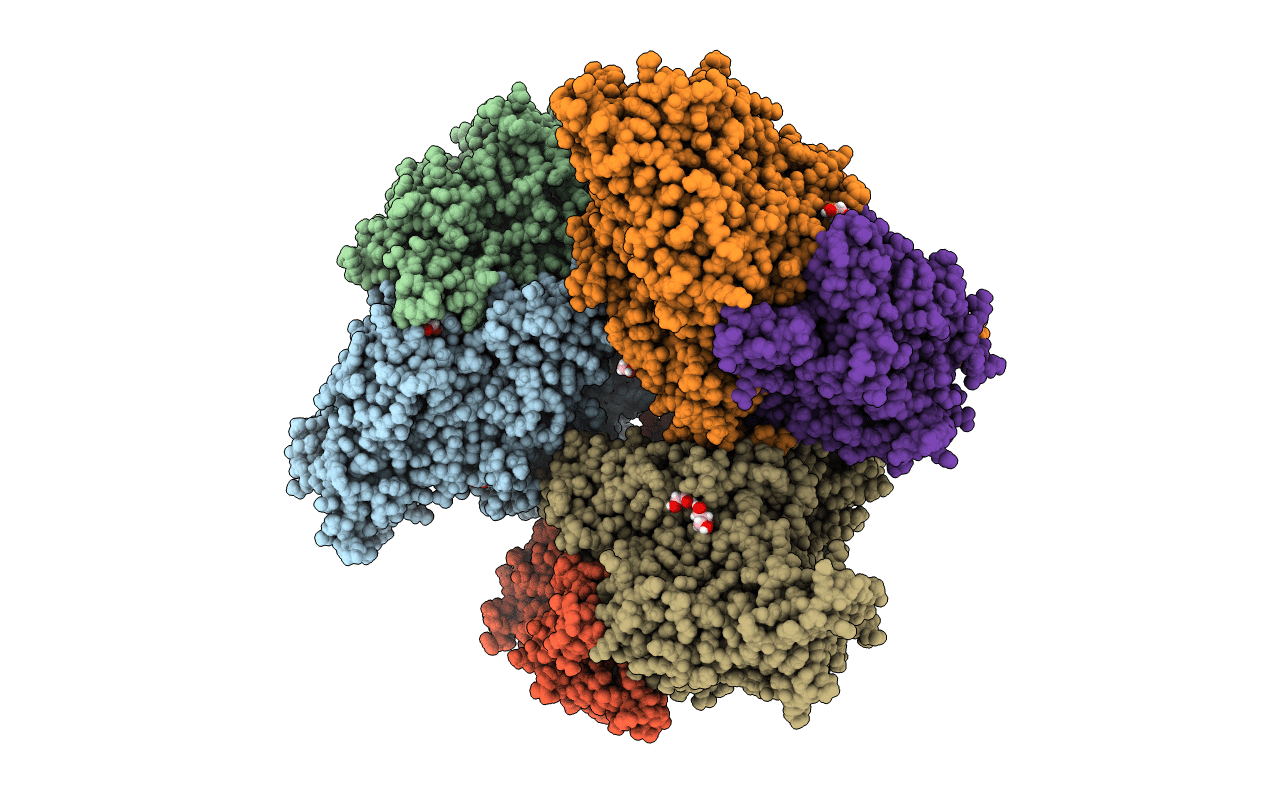 |
Crystal Structure Of The Perchlorate Reductase Pcrab From Azospira Suillum Ps
Organism: Dechlorosoma suillum
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, MD1, EDO, F3S, NA, GOL, PEG |
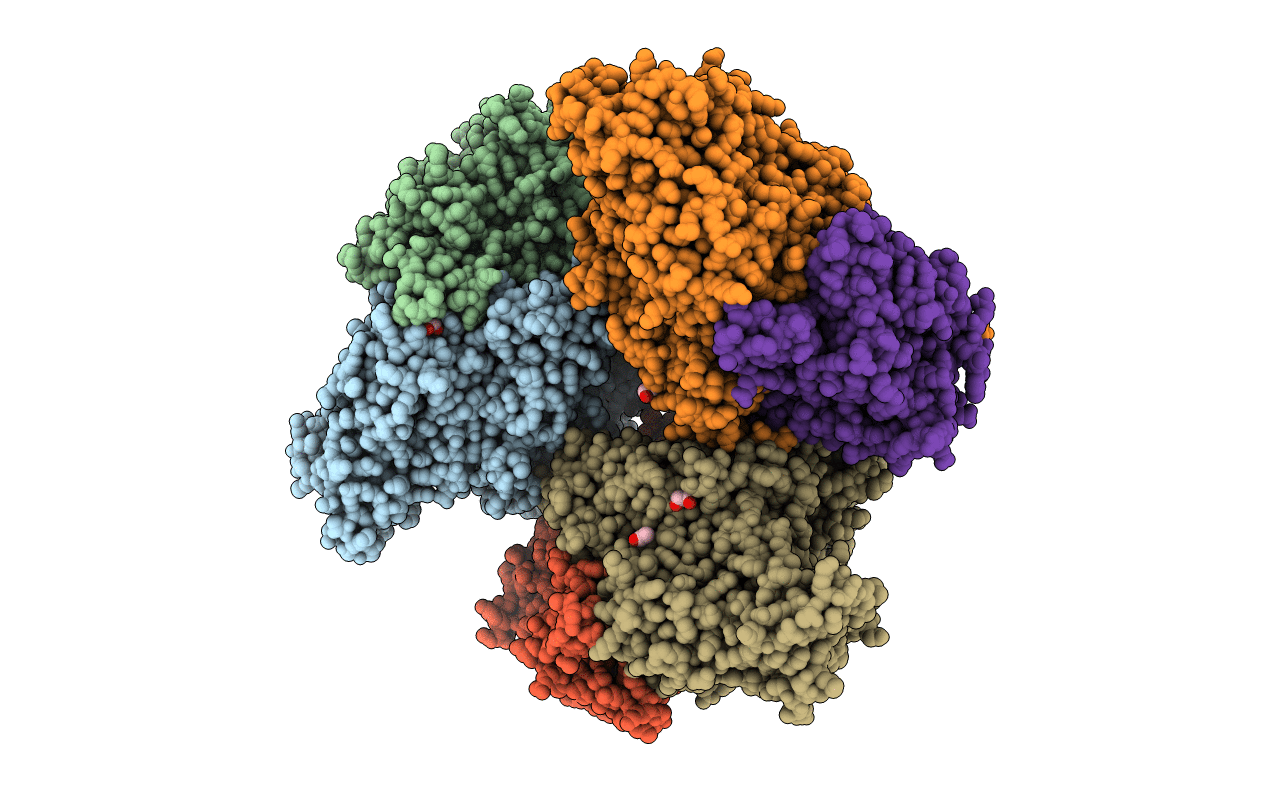 |
Crystal Structure Of The Perchlorate Reductase Pcrab - Phe164 Gate Switch Intermediate - From Azospira Suillum Ps
Organism: Azospira oryzae (strain atcc baa-33 / dsm 13638 / ps)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, MD1, EDO, NA, ZN, GOL, SO3, F3S, ACT |
 |
Crystal Structure Of The Perchlorate Reductase Pcrab - Substrate Analog Seo3 Bound - From Azospira Suillum Ps
Organism: Azospira oryzae (strain atcc baa-33 / dsm 13638 / ps)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, MD1, EDO, BSY, SO3, NA, ZN, F3S, GOL |
 |
Crystal Structure Of The Perchlorate Reductase Pcrab Mutant W461E Of Pcra From Azospira Suillum Ps
Organism: Dechlorosoma suillum (strain atcc baa-33 / dsm 13638 / ps)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE Ligands: SF4, MO, MGD, MD1, EDO, GOL, F3S |

