Search Count: 260
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH4 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH5 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH6 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH7 |
 |
Organism: Homo sapiens, Mus musculus, Hepatitis b virus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus, Hepatitis b virus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus, Hepatitis b virus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Lama glama, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: PEPTIDE BINDING PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of The Soluble Domain Of The Putative Ompa -Family Membrane Protein Ypo0514 From Yersinia Pestis
Organism: Yersinia pestis co92
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-01-26 Classification: TRANSPORT PROTEIN Ligands: CA, PO4 |
 |
Crystal Structure Of The Hiv Capsid Hexamer Bound To The Small Molecule Long-Acting Inhibitor, Gs-6207
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN Ligands: QNG |
 |
Human Pd-L1 Bound To A Macrocyclic Peptide Which Blocks The Pd-1/Pd-L1 Interaction
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-01-01 Classification: IMMUNE SYSTEM |
 |
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, FMT, EDO, CL, ZN, GOL |
 |
Crystal Structure Of Metallo Beta Lactamase From Hirschia Baltica In Complex With Succinate
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: ZN, SIN, EDO, FMT, BTB |
 |
Crystal Structure Of Metallo Beta Lactamase From Hirschia Baltica In The Complex With The Inhibitor Captopril
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: ZN, EDO, X8Z, NA, FMT, CL |
 |
Crystal Structure Of Metallo Beta Lactamase From Hirschia Baltica With Cadmium In The Active Site
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: CL, EDO, CD, FMT |
 |
Crystal Structure Of Metallo Beta Lactamase From Hirschia Baltica With Nitrate In The Active Site
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: ZN, EDO, NO3, FMT, PO4 |
 |
Organism: Erythrobacter litoralis (strain htcc2594)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: CA, CL, FMT, EDO |
 |
Crystal Structure Of Metallo Beta Lactamase From Erythrobacter Litoralis With Beta Mercaptoethanol In The Active Site
Organism: Erythrobacter litoralis (strain htcc2594)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: ZN, CL, BME, FMT |
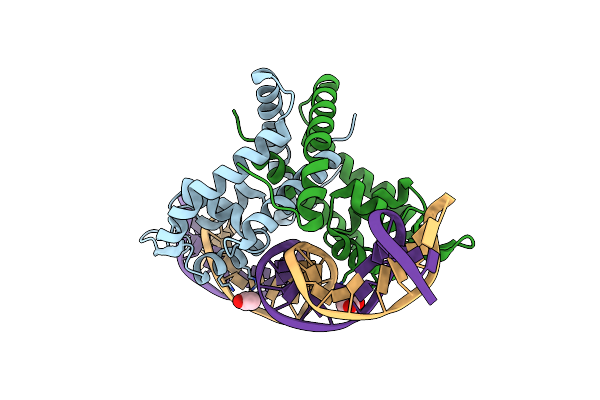 |
Crystal Structure Of Putative Marr Family Transcriptional Regulator From Listeria Monocytogenes Complexed With 26Mer Dna
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2019-08-21 Classification: TRANSCRIPTION Ligands: ACY |
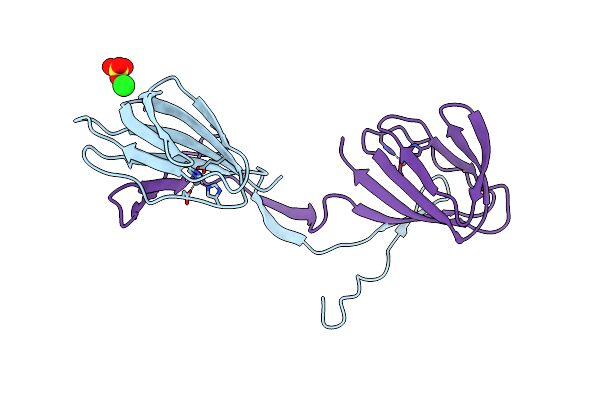 |
Organism: Rhodococcus jostii (strain rha1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-02-22 Classification: UNKNOWN FUNCTION Ligands: ZN, CL, SO4 |

