Search Count: 69
 |
Organism: Sporosarcina pasteurii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: HYDROLASE Ligands: NI, OH |
 |
Organism: Sporosarcina pasteurii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: HYDROLASE Ligands: NI, 2PA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-02-21 Classification: VIRAL PROTEIN |
 |
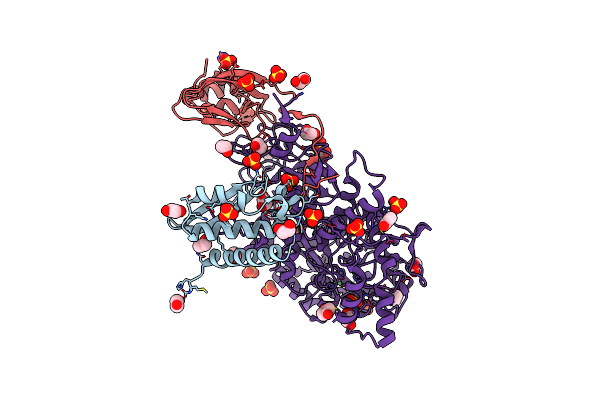
The 1.68-A X-Ray Crystal Structure Of Sporosarcina Pasteurii Urease Inhibited By Thiram And Bound To Dimethylditiocarbamate
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: EDO, SO4, IS9, NI, OH |
 |
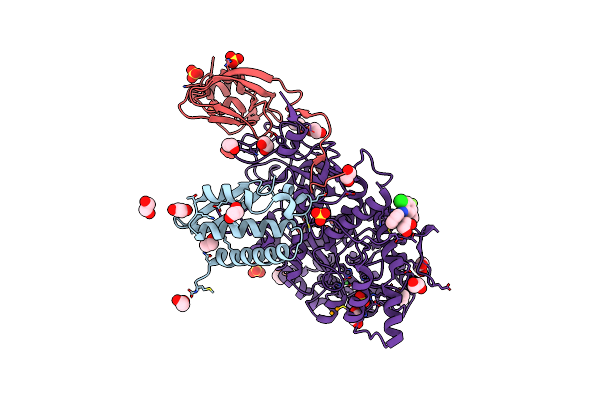
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: EDO, SO4, NI, OH, HQE |
 |
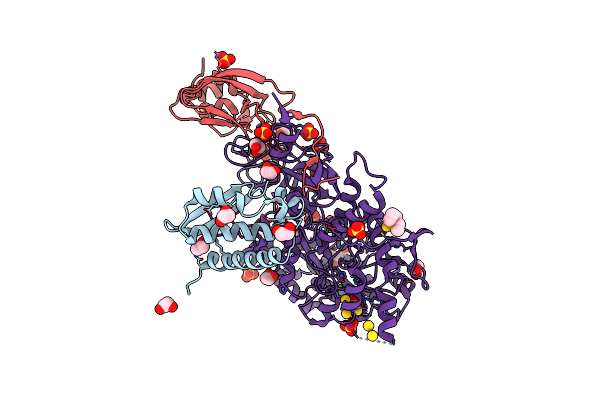
Sporosarcina Pasteurii Urease (Spu) Co-Crystallized In The Presence Of An Ebselen-Derivative And Bound To Se Atoms
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Release Date: 2023-04-19 Classification: HYDROLASE Ligands: EDO, SO4, NI, OH, IU9, SE |
 |
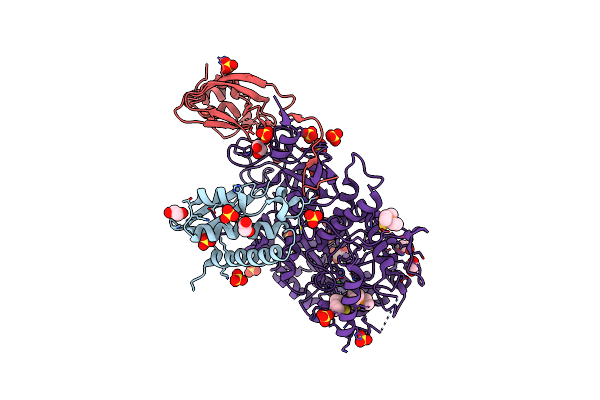
X-Ray Crystal Structure Of Sporosarcina Pasteurii Urease Inhibited By The Gold(I)-Phosphine Compound Au(Pet3)I Determined At 1.80 Angstroms
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: EDO, SO4, NI, O, AUF, AU |
 |
X-Ray Crystal Structure Of Sporosarcina Pasteurii Urease Inhibited By The Gold(I)-Diphosphine Compound Au(Pet3)2Cl Determined At 1.87 Angstroms
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: EDO, SO4, NI, O, AUF |
 |
1.93 A Resolution X-Ray Crystal Structure Of The Transcriptional Regulator Srnr From Streptomyces Griseus
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-05-25 Classification: DNA BINDING PROTEIN Ligands: ACT, NA |
 |
X-Ray Crystal Structure Of Sporosarcina Pasteurii Urease Inhibited By Ag(Pet3)Cl Determined At 1.72 Angstroms
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2021-10-13 Classification: HYDROLASE Ligands: EDO, SO4, NI, O, AG |
 |
X-Ray Crystal Structure Of Sporosarcina Pasteurii Urease Inhibited By Ag(Pet3)Br Determined At 1.63 Angstroms
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-10-13 Classification: HYDROLASE Ligands: EDO, SO4, NI, O, AG |
 |
X-Ray Crystal Structure Of Sporosarcina Pasteurii Urease Inhibited By Ag(Pet3)2No3 Determined At 1.97 Angstroms
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-10-13 Classification: HYDROLASE Ligands: EDO, SO4, NI, O, AG |
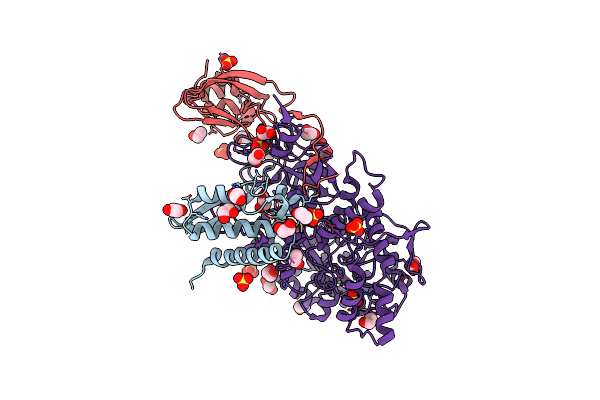 |
1.50 A Resolution 3-Methylcatechol (3-Methylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
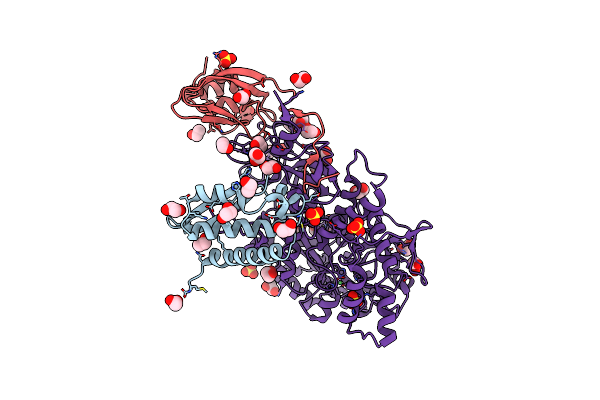 |
1.89 A Resolution 4-Methylcatechol (4-Methylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
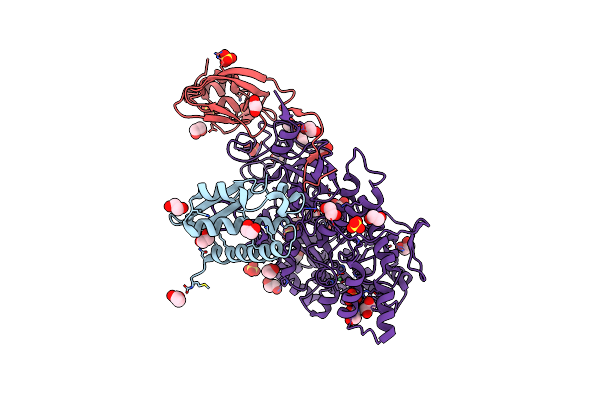 |
2.23 A Resolution 3,4-Dimethylcatechol (3,4-Dimethylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
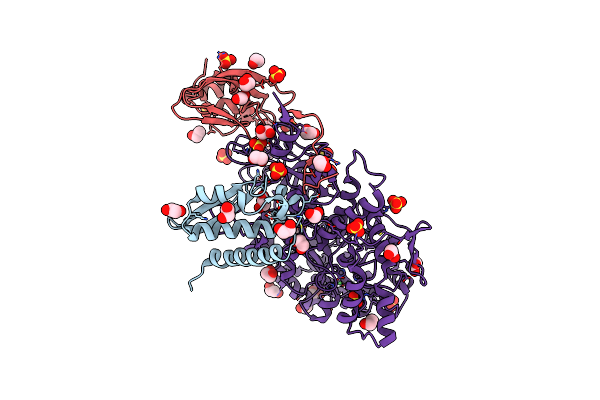 |
1.61 A Resolution 3,5-Dimethylcatechol (3,5-Dimethylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
 |
1.65 A Resolution 4,5-Dimethylcatechol (4,5-Dimethylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
 |
1.55 A Resolution 3,6-Dimethylcatechol (3,6-Dimethylbenzene-1,2-Diol) Inhibited Sporosarcina Pasteurii Urease
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-12-23 Classification: METAL BINDING PROTEIN Ligands: EDO, SO4, NI, OH |
 |
1.32 A Resolution Of Sporosarcina Pasteurii Urease Inhibited In The Presence Of Nbpto At Ph 7.5
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2020-03-11 Classification: HYDROLASE Ligands: EDO, SO4, NI, 2PA |
 |
1.49 A Resolution Of Sporosarcina Pasteurii Urease Inhibited In The Presence Of Nbpto At Ph 6.5
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2020-03-11 Classification: HYDROLASE Ligands: EDO, SO4, NI, 2PA |

