Search Count: 17
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: CU, ZN, ACT, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-03-06 Classification: HORMONE Ligands: IPH, ZN, CL, MYR |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NO3 |
 |
Tumor Necrosis Factor Receptor Associated Factor 6 (Traf6) N-Terminal Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-01-25 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Kitasatospora setae (strain atcc 33774 / dsm 43861 / jcm 3304 / kcc a-0304 / nbrc 14216 / km-6054)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: NAP, VES, 2EC, YAS, NDP |
 |
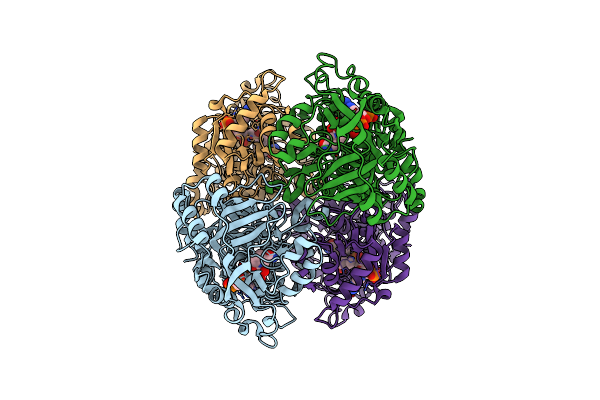
Organism: Kitasatospora setae (strain atcc 33774 / dsm 43861 / jcm 3304 / kcc a-0304 / nbrc 14216 / km-6054)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: VES, CL |
 |
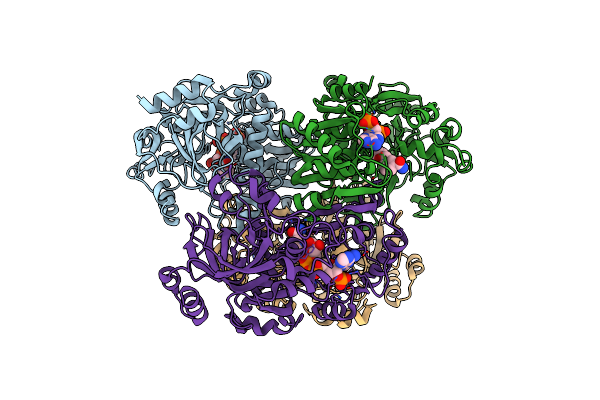
Organism: Kitasatospora setae (strain atcc 33774 / dsm 43861 / jcm 3304 / kcc a-0304 / nbrc 14216 / km-6054)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
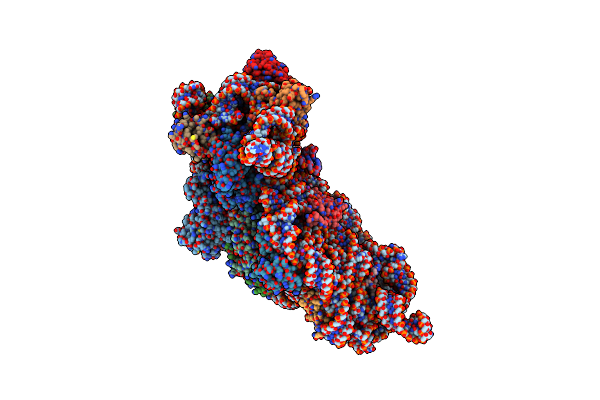
Serial Femtosecond X-Ray Crystallography Structure Of Ecr In Complex With Nadph
Organism: Kitasatospora setae (strain atcc 33774 / dsm 43861 / jcm 3304 / kcc a-0304 / nbrc 14216 / km-6054)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
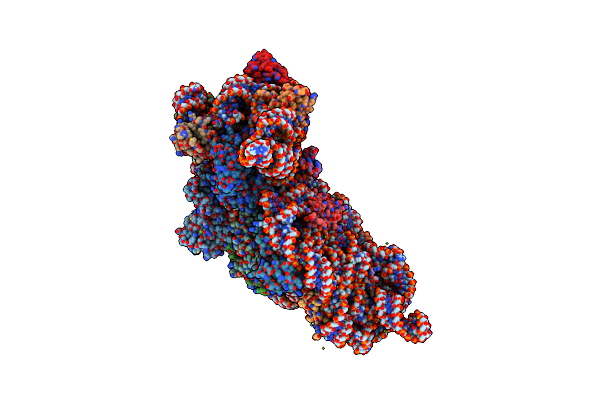
Organism: Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2018-07-25 Classification: RIBOSOME Ligands: PAR, K, MG, ZN |
 |
Serial Femtosecond X-Ray Crystal Structure Of 30S Ribosomal Subunit From Thermus Thermophilus In Complex With Sisomicin
Organism: Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-07-25 Classification: RIBOSOME Ligands: MG, SIS, ZN |
 |
Serial Femtosecond X-Ray Crystal Structure Of 30S Ribosomal Subunit From Thermus Thermophilus In Complex With N1Ms
Organism: Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2018-07-25 Classification: RIBOSOME Ligands: MG, EUS, ZN |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-16 Classification: Biotin binding protein Ligands: BYY |

