Search Count: 6
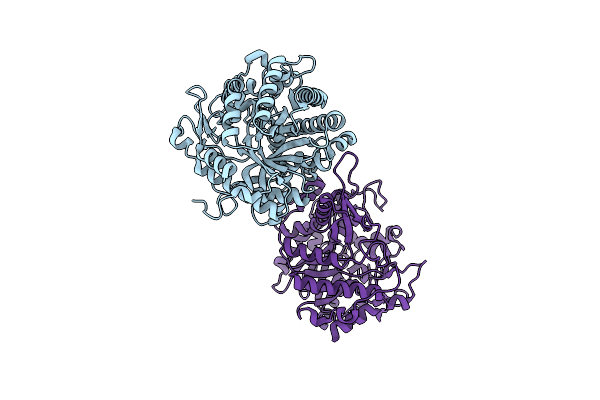 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2001-02-19 Classification: HYDROLASE |
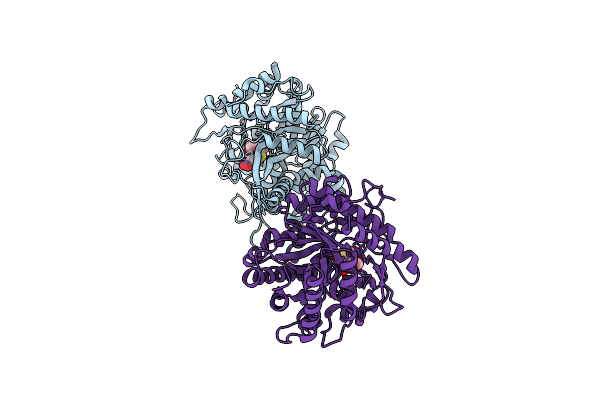 |
Crystal Structure Of A Monocot (Maize Zmglu1) Beta-Glucosidase In Complex With P-Nitrophenyl-Beta-D-Thioglucoside
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2001-02-19 Classification: HYDROLASE Ligands: PSG |
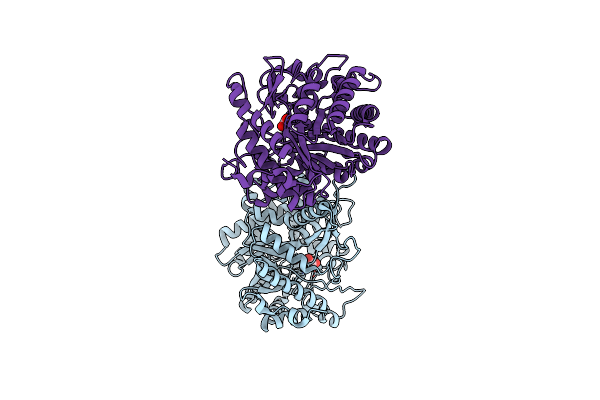 |
Crystal Structure Of The Inactive Mutant Monocot (Maize Zmglu1) Beta-Glucosidase Zm Glu191Asp
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2000-12-11 Classification: HYDROLASE Ligands: GOL |
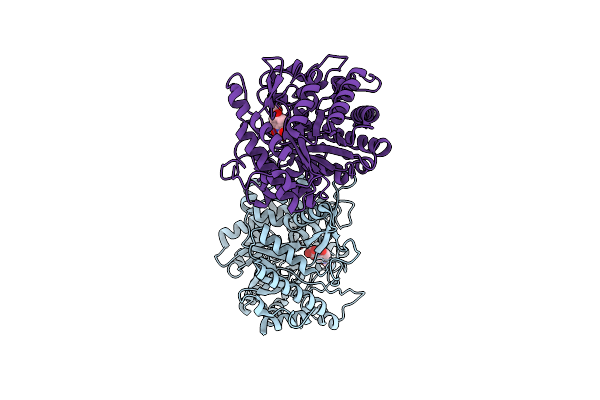 |
Crystal Structure Of The Inactive Mutant Monocot (Maize Zmglu1) Beta-Glucosidase Zmglue191D In Complex With The Natural Aglycone Dimboa
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-12-11 Classification: HYDROLASE Ligands: HBO |
 |
Crystal Structure Of The Inactive Mutant Monocot (Maize Zmglu1) Beta-Glucosidase Zmglue191D In Complex With The Competitive Inhibitor Dhurrin
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-12-11 Classification: HYDROLASE Ligands: BGC, DHR |
 |
Crystal Structure Of The Inactive Mutant Monocot (Maize Zmglu1) Beta-Glucosidase Zmglue191D In Complex With The Natural Substrate Dimboa-Beta-D-Glucoside
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-12-11 Classification: HYDROLASE Ligands: BGC, HBO |

