Search Count: 21
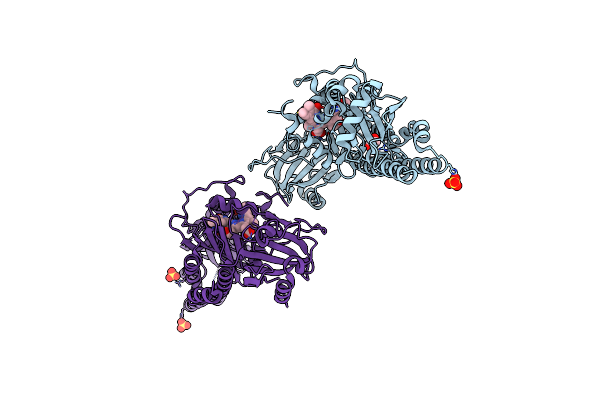 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
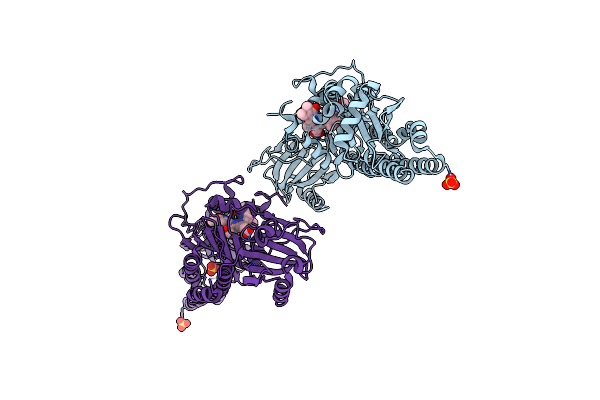 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
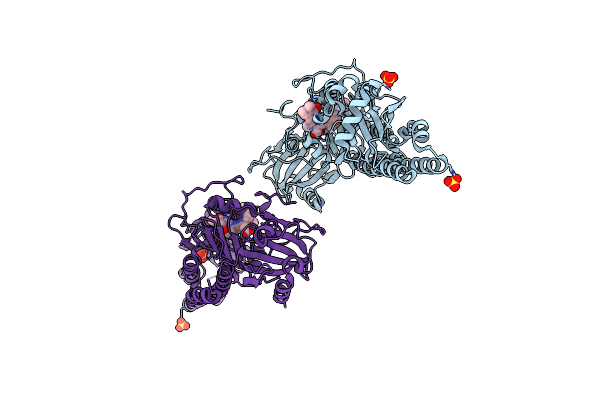 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
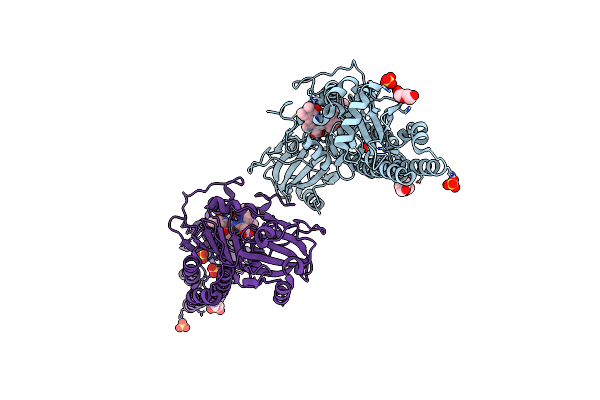 |
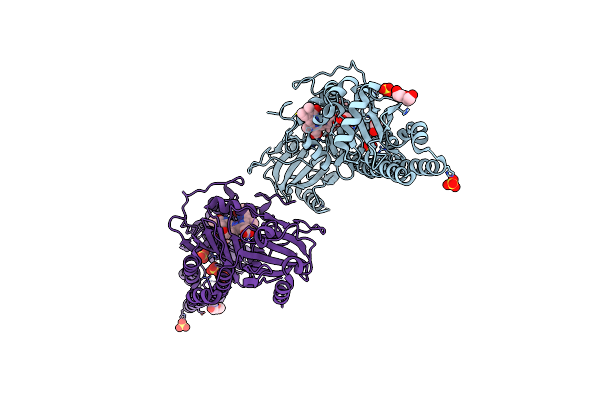
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
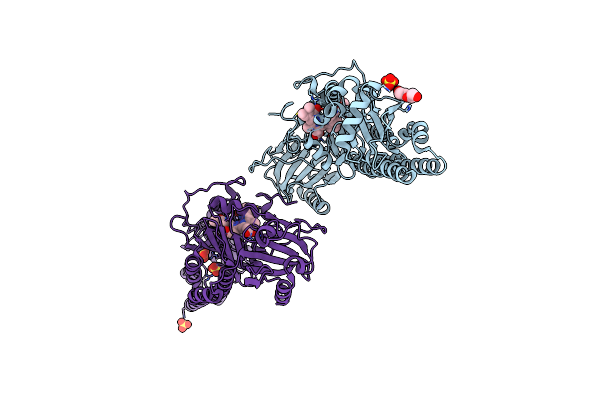
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
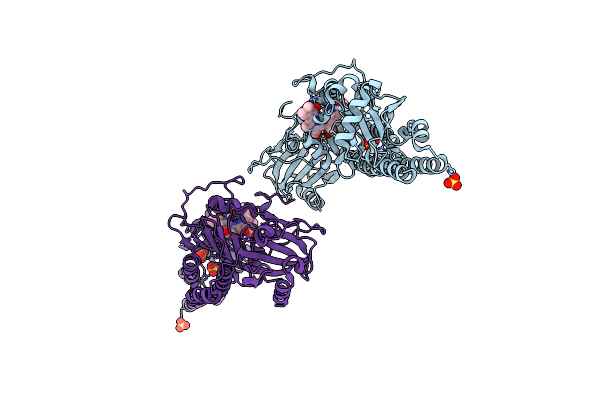
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
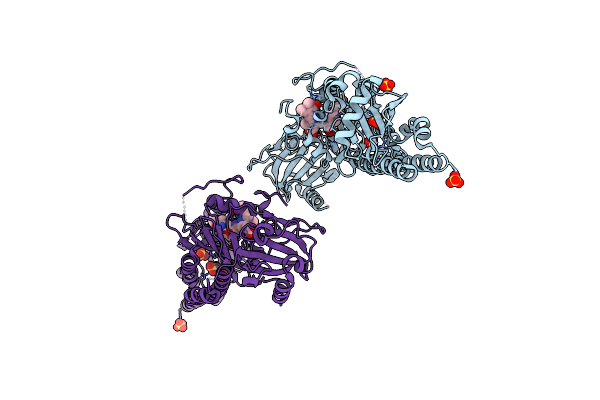
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-18 Classification: FLUORESCENT PROTEIN |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2023-10-18 Classification: FLUORESCENT PROTEIN |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-09-27 Classification: FLUORESCENT PROTEIN |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2023-07-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, SO4 |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2023-07-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, SO4 |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-07-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, SO4 |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2022-11-09 Classification: FLUORESCENT PROTEIN Ligands: SO4 |
 |
Organism: Synechocystis
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2022-11-09 Classification: PHOTOSYNTHESIS Ligands: 45D, GOL, ACT, CL |
 |
Structure Of Orange Carotenoid Protein With Canthaxanthin Bound After 1 Minute Of Illumination
Organism: Synechocystis
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2022-11-09 Classification: PHOTOSYNTHESIS Ligands: 45D, GOL, ACT, CL |
 |
Structure Of Orange Carotenoid Protein With Canthaxanthin Bound After 2 Minutes Of Illumination
Organism: Synechocystis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2022-11-09 Classification: PHOTOSYNTHESIS Ligands: 45D, GOL, ACT, CL |
 |
Structure Of Orange Carotenoid Protein With Canthaxanthin Bound After 5 Minutes Of Illumination
Organism: Synechocystis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-11-09 Classification: PHOTOSYNTHESIS Ligands: 45D, GOL, ACT, CL |

