Search Count: 8
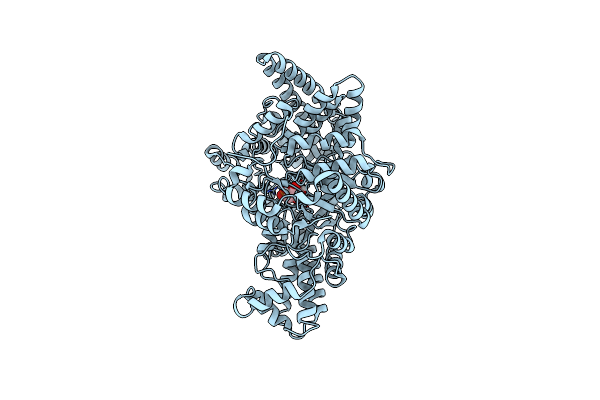 |
Crystal Structure Of Pfld Bound To 1,5-Anhydromannitol-6-Phosphate In Streptococcus Dysgalactiae Subsp. Equisimilis
Organism: Streptococcus dysgalactiae subsp. equisimilis
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 7P6 |
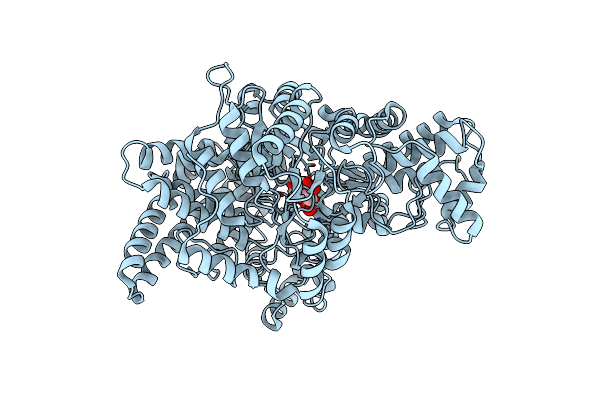 |
Crystal Structure Of Ybiw In Complex With 1,5-Anhydroglucitol-6-Phosphate In Escherichia Coli
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 0WK |
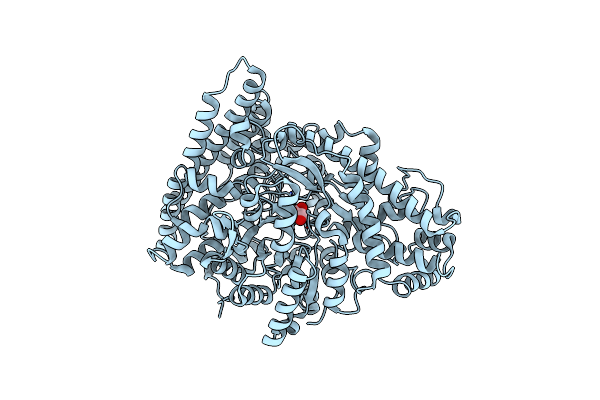 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-10-02 Classification: LYASE Ligands: GOL |
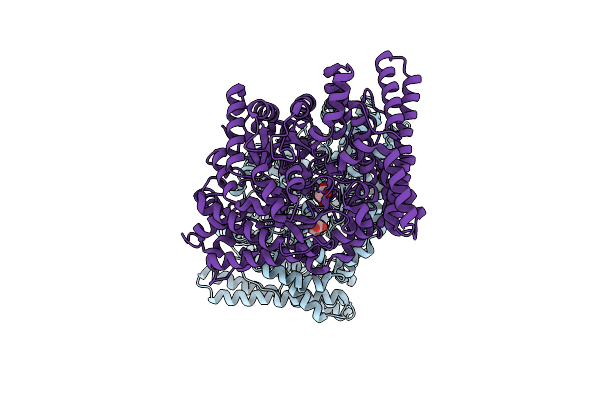 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-02 Classification: LYASE Ligands: MLI |
 |
Crystal Structure Of Macrolide Phosphotransferase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2024-01-31 Classification: TRANSFERASE |
 |
Organism: Rhodococcus wratislaviensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: ZN, DPO, GLY |
 |
Organism: Rhodococcus wratislaviensis
Method: X-RAY DIFFRACTION Release Date: 2021-07-28 Classification: HYDROLASE Ligands: DUT, ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2002-11-06 Classification: ELECTRON TRANSPORT Ligands: SO4 |

