Search Count: 926
All
Selected
 |
Crystal Structure Of Cim-2, A Membrane-Bound B1 Metallo-Beta-Lactamase From Chryseobacterium Indologenes
Organism: Chryseobacterium indologenes
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-07-09 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN, MPD, CL, IPA, PGR |
 |
Crystal Structure Of Cjo-1, A Membrane-Bound B1 Metallo-Beta-Lactamase From Chryseobacterium Joostei
Organism: Chryseobacterium joostei
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-07-09 Classification: ANTIMICROBIAL PROTEIN Ligands: GOL, ZN |
 |
Organism: Chryseobacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: DNA BINDING PROTEIN/DNA/RNA |
 |
Organism: Chryseobacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: DNA BINDING PROTEIN/DNA/RNA |
 |
Structure Of Cbcas9-Pcriic1 Complex Bound To 28-Bp Dna Substrate (20-Nt Complementary)
Organism: Chryseobacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Structure Of Cbcas9-Pcriic1 Complex Bound To 62-Bp Dna Substrate (Symmetric 20-Nt Complementary)
Organism: Chryseobacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Structure Of Cbcas9-Pcriic1 Complex Bound To 62-Bp Dna Substrate (Non-Targeting Complex)
Organism: Chryseobacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Organism: Chryseobacterium oncorhynchi
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-04-26 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: TDC, FAD |
 |
Organism: Chryseobacterium oncorhynchi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-04-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Cytoplasmic Domain Structure Of The Mgte Mg2+ Channel From Chryseobacterium Hispalense
Organism: Chryseobacterium hispalense
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2023-04-19 Classification: TRANSPORT PROTEIN Ligands: MG |
 |
Organism: Chryseobacterium sp. yr480
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-04-13 Classification: HYDROLASE Ligands: CL |
 |
Organism: Chryseobacterium jeonii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: K |
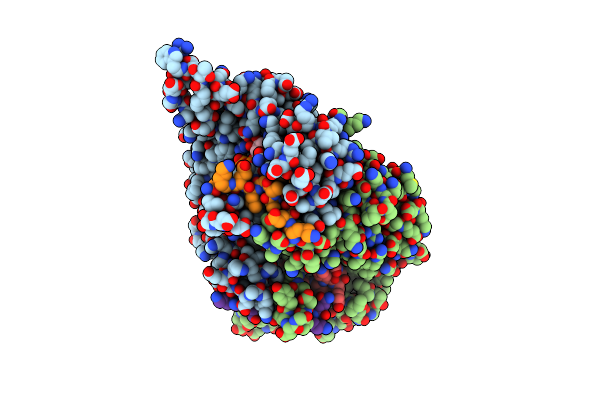 |
Chryseobacterium Gregarium Ripp-Associated Atp-Grasp Ligase In Complex With Adp, And A Leader And Core Peptide
Organism: Chryseobacterium gregarium dsm 19109
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2021-06-23 Classification: LIGASE Ligands: ADP, GOL |
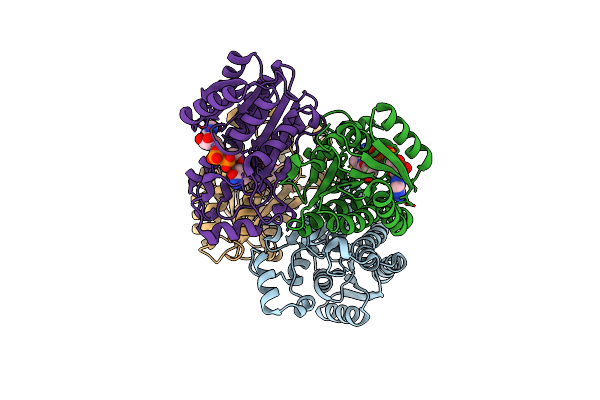 |
Crystal Structure Of The Ketone Reductase Chkred20 From The Genome Of Chryseobacterium Sp. Ca49 Complexed With Nad
Organism: Chryseobacterium sp. ca49
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-04-03 Classification: OXIDOREDUCTASE Ligands: NAD |
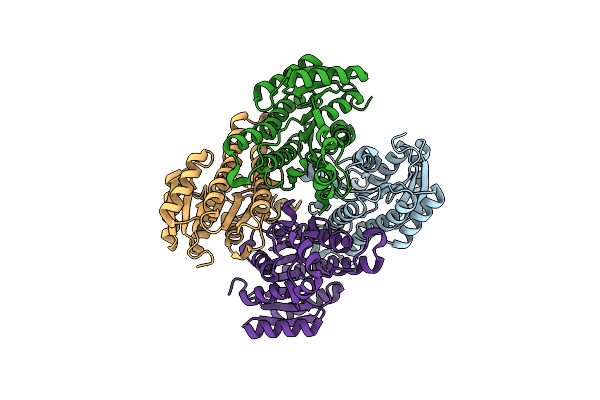 |
Crystal Structure Of The Ketone Reductase Chkred20 From The Genome Of Chryseobacterium Sp. Ca49
Organism: Chryseobacterium sp. ca49
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-11-08 Classification: OXIDOREDUCTASE |
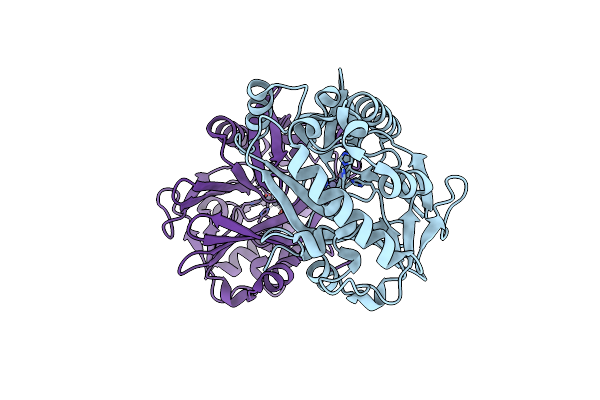 |
Organism: Chryseobacterium sp. strb126
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: ZN |
 |
Aidc, A Dizinc Quorum-Quenching Lactonase, In Complex With A Product N-Hexnoyl-L-Homoserine
Organism: Chryseobacterium sp. strb126
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: ZN, C6L |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-08-11 Classification: HYDROLASE |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-08-11 Classification: HYDROLASE |
 |
Organism: Chryseobacterium proteolyticum
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2010-08-11 Classification: HYDROLASE Ligands: CIT |

