Search Count: 27
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: AN6 |
 |
Organism: Vibrio cholerae, Vibrio cholerae o1
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/DNA Ligands: AGS, MG |
 |
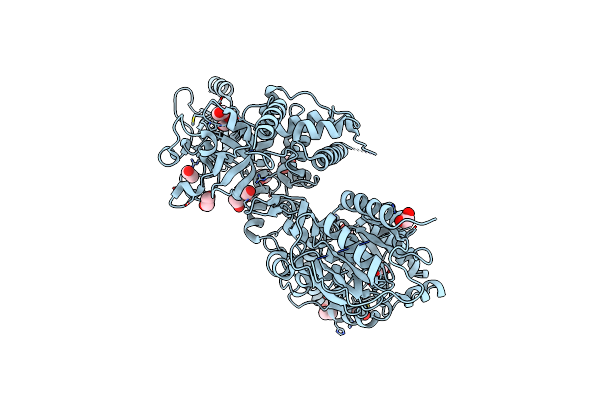
Structure Of The Smurf2 Hect Domain With A High Affinity Ubiquitin Variant (Ubv)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-21 Classification: SIGNALING PROTEIN/TRANSFERASE Ligands: SO4, NA, CL, DTT, EDO |
 |
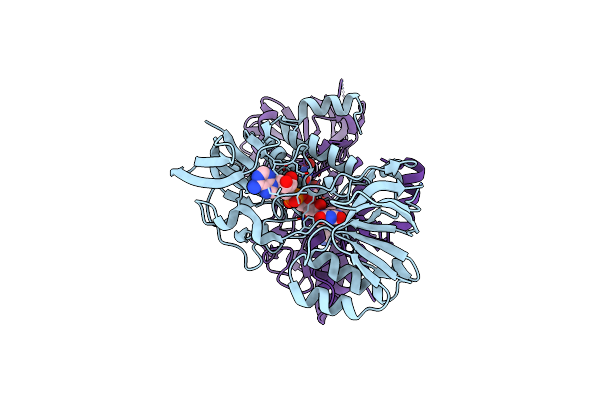
Crystal Structure Of S. Cerevisiae Deah-Box Rna Helicase Dhr1, Essential For Small Ribosomal Subunit Biogenesis
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-06-19 Classification: HYDROLASE Ligands: EDO, MG, CL |
 |
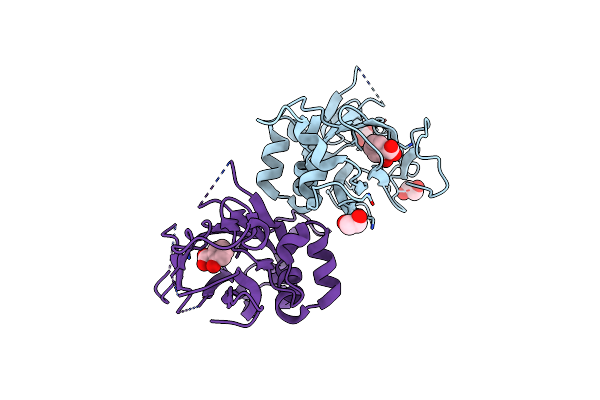
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-07-11 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
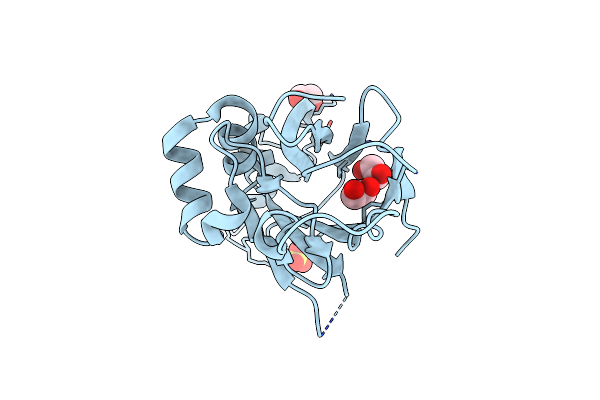
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2018-03-21 Classification: PROTEIN FIBRIL Ligands: SAL, EDO |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-03-21 Classification: PROTEIN FIBRIL Ligands: EDO, SO4 |
 |
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2015-12-09 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Post-Catalytic Binary Complex Of Phosphoglycerate Mutase From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-05-06 Classification: ISOMERASE Ligands: MN, 2PG |
 |
Crystal Structure Of Phosphopglycerate Mutase From Staphylococcus Aureus In 3-Phosphoglyceric Acid Bound Form.
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-01-14 Classification: ISOMERASE Ligands: MN, 3PG |
 |
Crystal Structure Of Phosphoglycerate Mutase From Staphylococcus Aureus In 2-Phosphoglyceric Acid Bound Form
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-01-14 Classification: ISOMERASE Ligands: 2PG, MN, DTT |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-10-16 Classification: ISOMERASE Ligands: MN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2013-08-07 Classification: RNA Ligands: SAM, MG |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-01-30 Classification: TRANSFERASE |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2013-01-02 Classification: OXIDOREDUCTASE Ligands: NAD, PO4, DGY |
 |
Crystal Structure Of A Beta-Ketoacyl Reductase Fabg4 From Mycobacterium Tuberculosis H37Rv Complexed With Nad+ And Hexanoyl-Coa At 2.5 Angstrom Resolution
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-11-28 Classification: OXIDOREDUCTASE Ligands: NAD, HXC, ZPG |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2012-11-28 Classification: OXIDOREDUCTASE Ligands: NAI, ZPG |
 |
Crystal Structure Of Staphylococcus Aureus Triosephosphate Isomerase Complexed With Glycerol-3-Phosphate
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-10-17 Classification: ISOMERASE Ligands: G3P, CIT |

