Search Count: 20
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-07-22 Classification: HYDROLASE |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2015-07-22 Classification: HYDROLASE Ligands: IOD |
 |
A Triple Mutant In The Omega-Loop Of Tem-1 Beta-Lactamase Changes The Substrate Profile Via A Large Conformational Change And An Altered General Base For Deacylation
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: BCT |
 |
A Triple Mutant In The Omega-Loop Of Tem-1 Beta-Lactamase Changes The Substrate Profile Via A Large Conformational Change And An Altered General Base For Catalysis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: SO4 |
 |
A Triple Mutant In The Omega-Loop Of Tem-1 Beta-Lactamase Changes The Substrate Profile Via A Large Conformational Change And An Altered General Base For Catalysis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: FLC |
 |
Crystallographic Analysis Of Rotavirus Nsp2-Rna Complex Reveals Specific Recognition Of 5'-Gg Sequence For Rtpase Activity
Organism: Simian 11 rotavirus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-08-22 Classification: HYDROLASE/RNA Ligands: CL, NA |
 |
Crystallographic Analysis Of Rotavirus Nsp2-Rna Complex Reveals Specific Recognition Of 5'-Gg Sequence For Rtpase Activity
Organism: Simian 11 rotavirus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2012-08-22 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2011-10-12 Classification: CHAPERONE |
 |
Structural, Thermodynamic And Kinetic Analysis Of The Picomolar Binding Affinity Interaction Of The Beta-Lactamase Inhibitor Protein-Ii (Blip-Ii) With Class A Beta-Lactamases
Organism: Bacillus anthracis, Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2011-07-20 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Structural, Thermodynamic And Kinetic Analysis Of The Picomolar Binding Affinity Interaction Of The Beta-Lactamase Inhibitor Protein-Ii (Blip-Ii) With Class A Beta-Lactamases
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-07-20 Classification: HYDROLASE INHIBITOR Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2011-05-11 Classification: TRANSFERASE Ligands: CMP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2011-05-11 Classification: TRANSFERASE Ligands: PCG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2011-05-11 Classification: TRANSFERASE Ligands: CMP, PO4 |
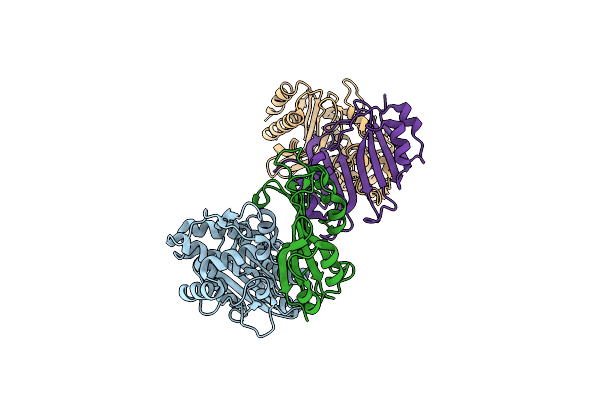 |
Structural Insight Into The Kinetics And Cp Of Interactions Between Tem-1-Lactamase And Blip
Organism: Escherichia coli, Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-10-07 Classification: HYDROLASE/HYDROLASE INHIBITOR |
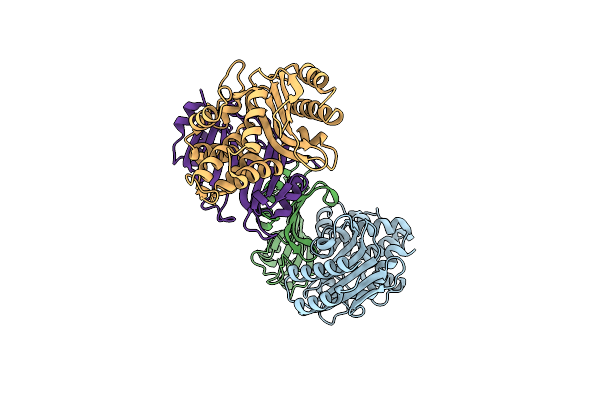 |
Structural Insight Into The Kinetics And Delta-Cp Of Interactions Between Tem-1 Beta-Lactamase And Blip
Organism: Escherichia coli, Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2008-10-07 Classification: HYDROLASE/HYDROLASE INHIBITOR |
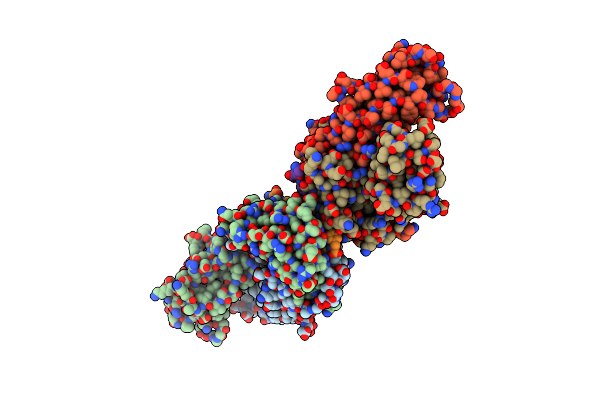 |
Evidence That Structural Rearrangements And/Or Flexibility During Tcr Binding Can Contribute To T-Cell Activation
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2004-03-02 Classification: SIGNALING PROTEIN |
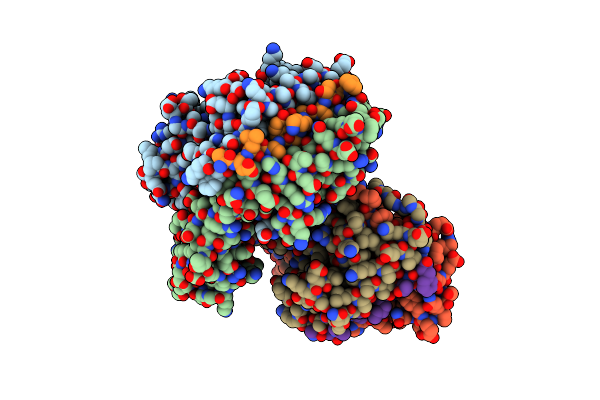 |
Evidence That Structural Rearrangements And/Or Flexibility During Tcr Binding Can Contribute To T-Cell Activation
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-02-03 Classification: SIGNALING PROTEIN |
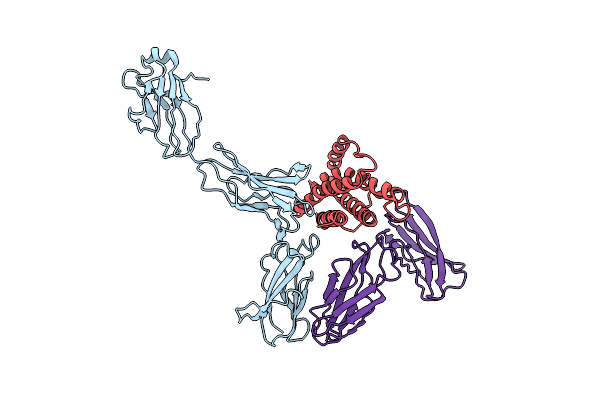 |
Crystal Structure Of The Hexameric Human Il-6/Il-6 Alpha Receptor/Gp130 Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2003-07-01 Classification: signaling protein/cytokine |
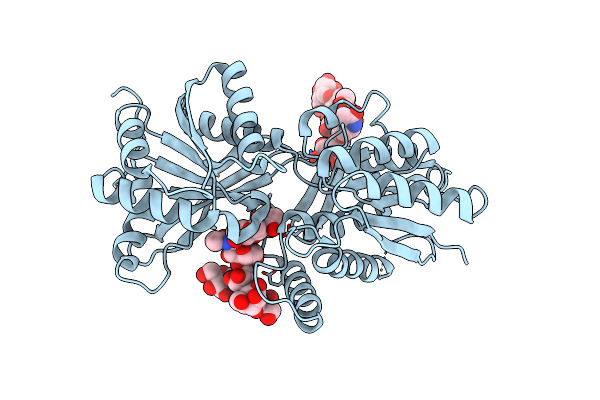 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2001-09-05 Classification: SIGNALING PROTEIN Ligands: CL |
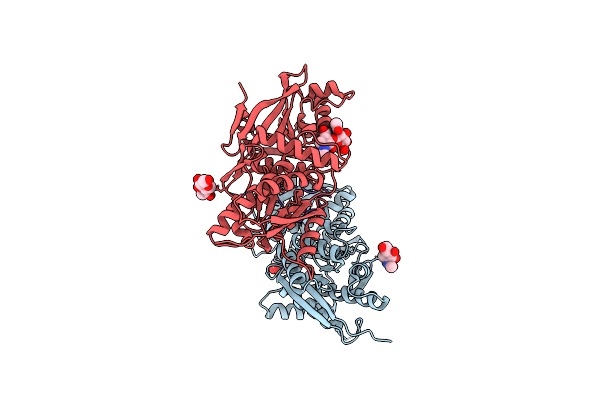 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-09-05 Classification: SIGNALING PROTEIN Ligands: NAG, CL |

