Search Count: 18
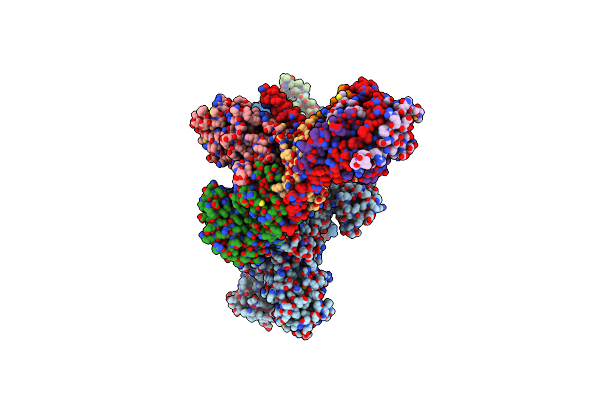 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: HYDROLASE Ligands: GNP |
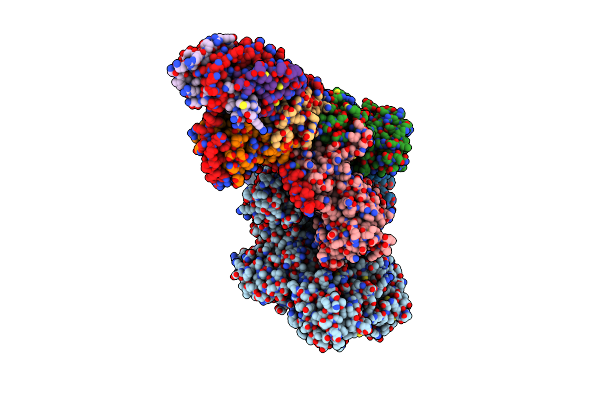 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: HYDROLASE Ligands: GDP, AF3 |
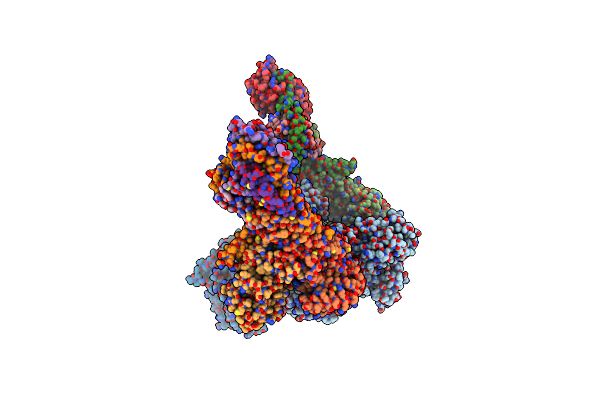 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: HYDROLASE Ligands: GDP, AF3 |
 |
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: PLANT PROTEIN, Lyase |
 |
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: PLANT PROTEIN |
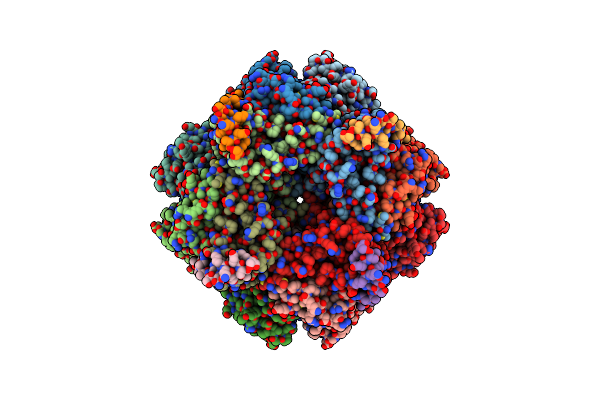 |
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: PLANT PROTEIN, Lyase |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2020-10-14 Classification: MEMBRANE PROTEIN Ligands: SO4, IOD |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-10-14 Classification: MEMBRANE PROTEIN Ligands: SCN |
 |
Organism: Staphylococcus aureus, Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2020-10-14 Classification: MEMBRANE PROTEIN Ligands: AGS, MG |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2020-10-14 Classification: MEMBRANE PROTEIN Ligands: AGS, MG |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: PROTEIN TRANSPORT |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2020-06-24 Classification: PROTEIN TRANSPORT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-11-20 Classification: SIGNALING PROTEIN Ligands: GDP, GNP, MG |
 |
Crystal Structure Of Native Heptameric Sap-Like Pentraxin From Limulus Polyphemus
Organism: Limulus polyphemus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-01-20 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of Native Octameric Sap-Like Pentraxin From Limulus Polyphemus
Organism: Limulus polyphemus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-01-20 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of Pe-Bound Octameric Sap-Like Pentraxin From Limulus Polyphemus
Organism: Limulus polyphemus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-01-20 Classification: SUGAR BINDING PROTEIN Ligands: CA, OPE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-11-14 Classification: TRANSFERASE Ligands: XUL, SO4, NH4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-11-14 Classification: TRANSFERASE |

