Search Count: 29
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In A P1 Crystal Form
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: LMR, IMD, GOL |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In A C2 Crystal Form
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: CIT, CO3 |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In Complex With Products
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: IMD, GOL, 98X |
 |
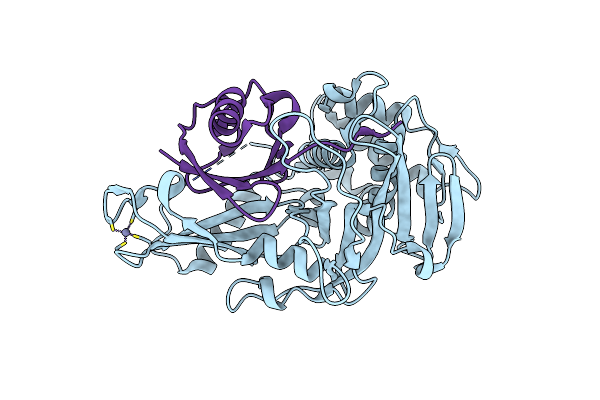
Porcine Epidemic Diarrhea Virus Papain-Like Protease 2 C44S Mutant In Complex With Mono Ubiquitin
Organism: Porcine epidemic diarrhea virus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-10-20 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: ZN |
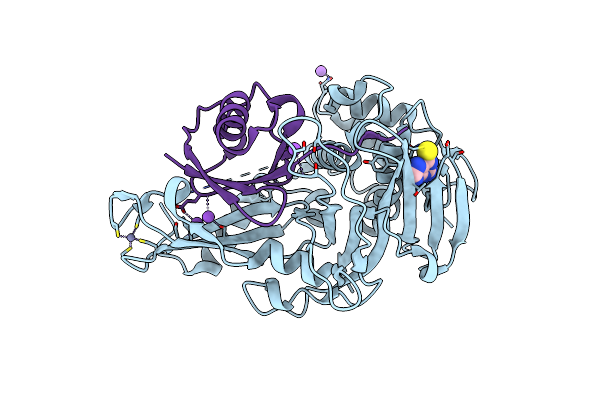 |
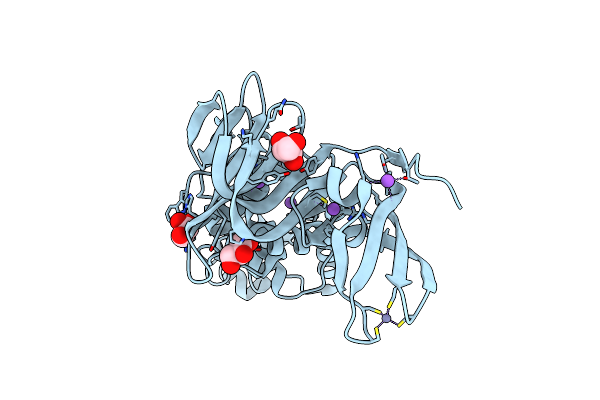
Crystal Structure Of Human Usp2 In Complex With Ubiquitin And 6-Thioguanine
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2018-02-28 Classification: HYDROLASE Ligands: DX4, ZN, CL, NA |
 |
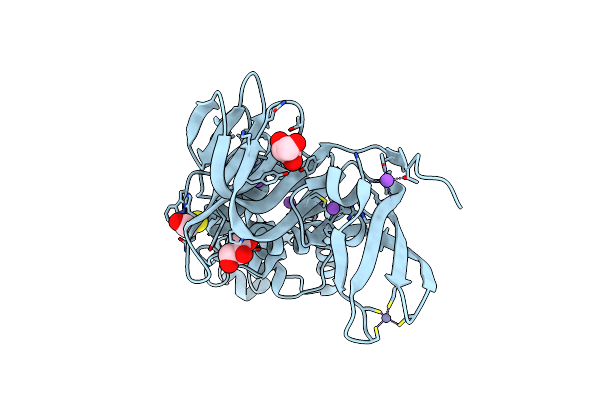
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2018-02-28 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Sars Coronavirus Papain-Like Protease In Complex With Glycerol
Organism: Human sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-01-10 Classification: HYDROLASE Ligands: ZN, NA, GOL |
 |
Crystal Structure Of Sars Coronavirus Papain-Like Protease Conjugated With Beta-Mercaptoethanol
Organism: Human sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-01-10 Classification: HYDROLASE Ligands: ZN, BME, NA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2017-11-15 Classification: GENE REGULATION |
 |
Crystal Structure Of Human Flash N-Terminal Domain C54S/C83A (Crystal Form 1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-11-15 Classification: GENE REGULATION Ligands: EDO |
 |
Crystal Structure Of Human Flash N-Terminal Domain C54S/C83A (Crystal Form 2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2017-11-15 Classification: GENE REGULATION |
 |
Crystal Structure Of Two-Subunit Pyruvate Carboxylase From Methylobacillus Flagellatus
Organism: Methylobacillus flagellatus, Methylobacillus flagellatus (strain kt / atcc 51484 / dsm 6875)
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2016-10-19 Classification: LIGASE Ligands: BTI, PYR, MN |
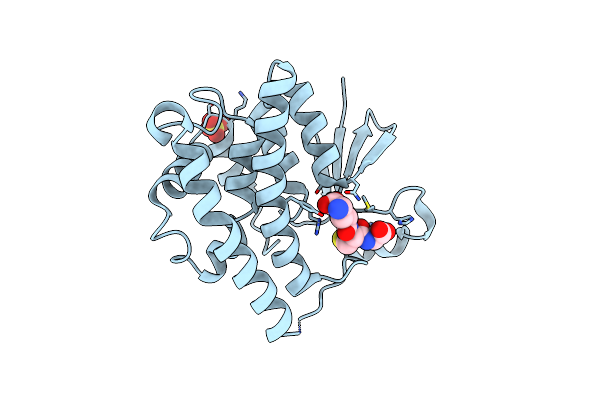 |
Crystal Structure Of Octopus S-Crystallin Q108F Mutant In Complex With Glutathione
Organism: Octopus vulgaris
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-08-03 Classification: STRUCTURAL PROTEIN Ligands: GSH, SO4 |
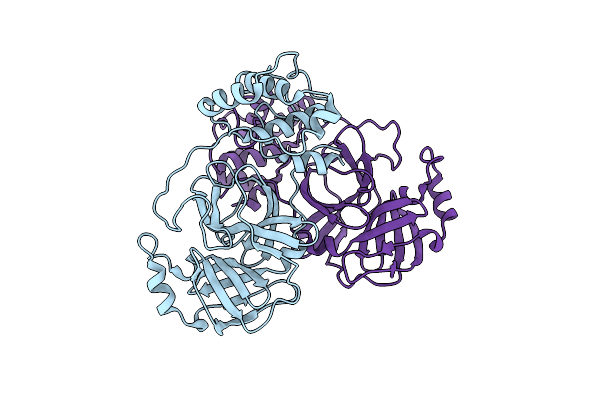 |
Organism: Middle east respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-12-30 Classification: HYDROLASE |
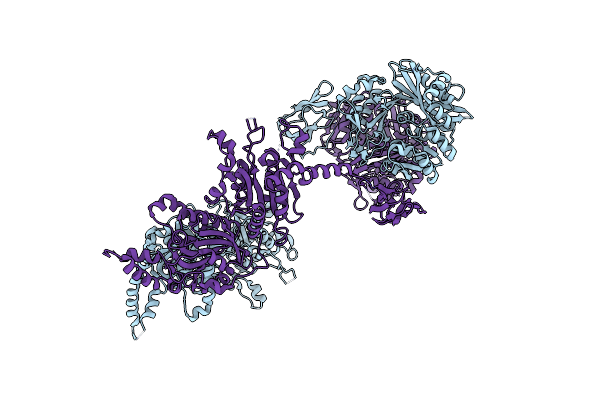 |
Structure And Function Of A Single-Chain, Multi-Domain Long-Chain Acyl-Coa Carboxylase
Organism: Mycobacterium avium subsp. paratuberculosis
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2014-11-05 Classification: LIGASE |
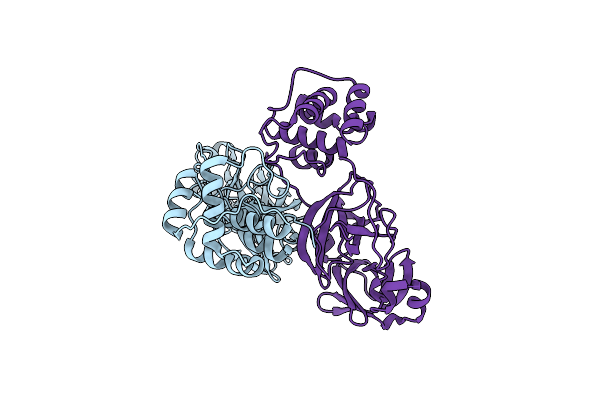 |
Crystal Structure Of Dimeric R298A Mutant Of Sars Coronavirus Main Protease
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2013-05-01 Classification: HYDROLASE |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-01-16 Classification: LIGASE Ligands: ADP, MN, BTI, ATP |
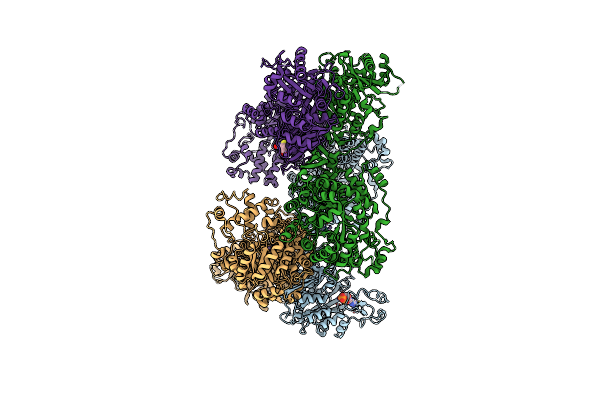 |
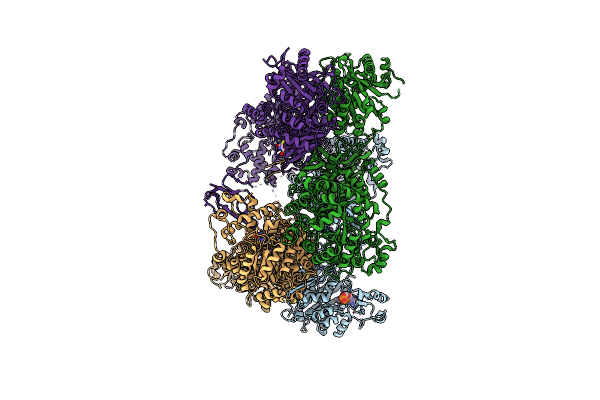
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-01-16 Classification: LIGASE Ligands: ADP, MN, BTI |
 |
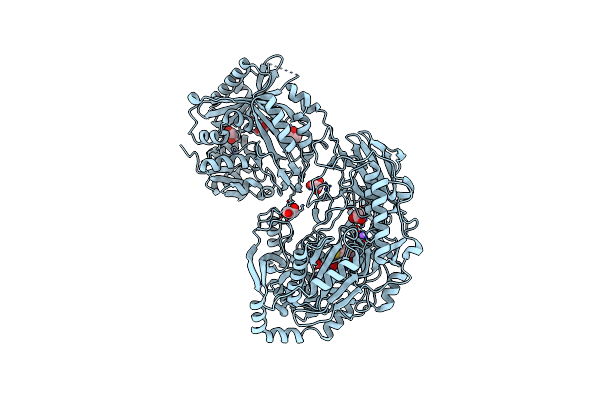
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-01-16 Classification: LIGASE Ligands: ADP, MN, BTI, ATP, CL |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-02-01 Classification: LIGASE Ligands: BTI, URE, GOL, NA |

