Search Count: 22
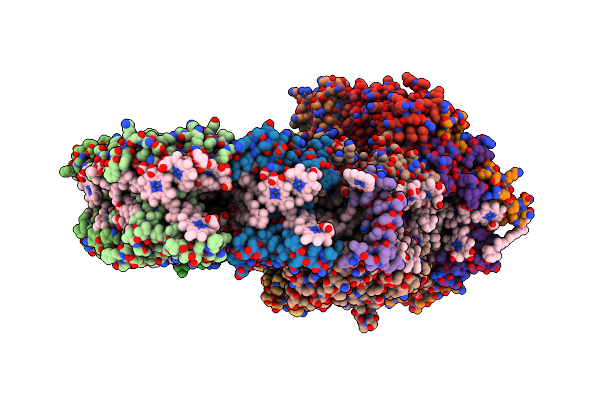 |
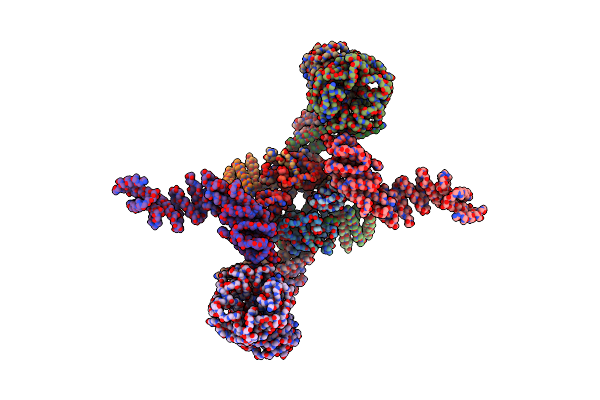
X-Ray Structure Of Photosystem I-Lhci Super Complex From Chlamydomonas Reinhardtii.
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-06-14 Classification: PHOTOSYNTHESIS Ligands: CLA, SF4, PQN, LHG, BCR, DGD, LMT, LMG, LUT, CHL, XAT |
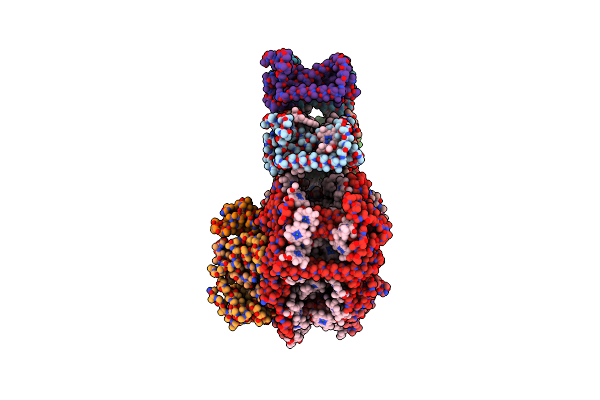 |
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Resolution:3.90 Å Release Date: 2023-02-22 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, CHL |
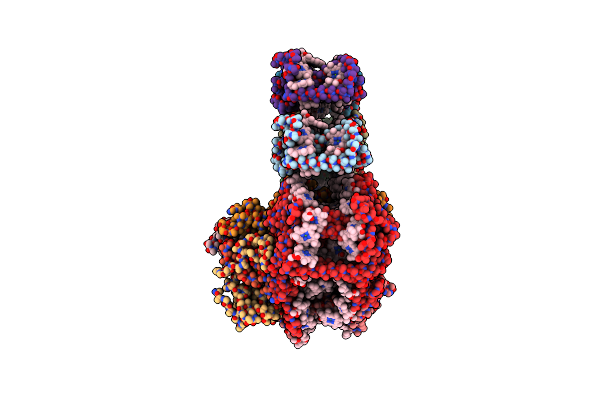 |
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Resolution:4.90 Å Release Date: 2023-02-22 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, CHL, FES |
 |
 |
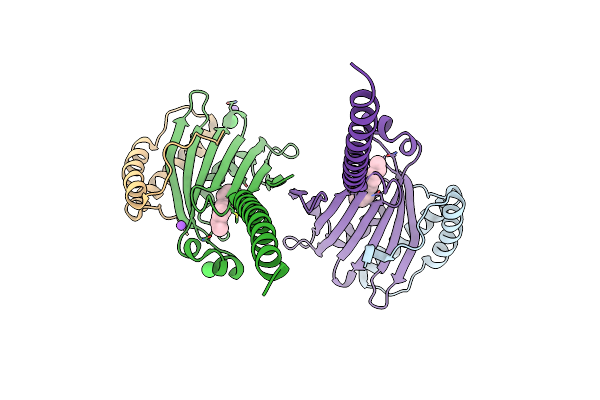 |
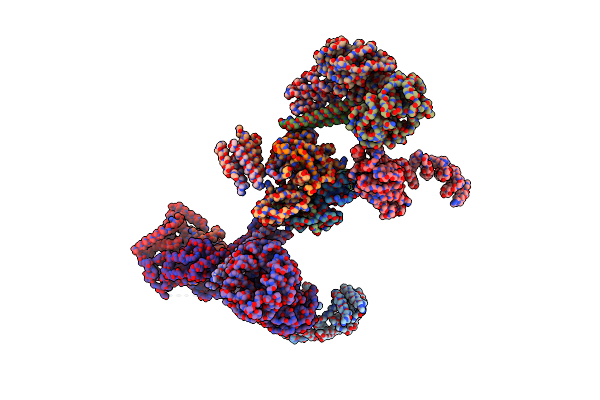
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-03-20 Classification: LIPID BINDING PROTEIN Ligands: MYR, CL, NA |
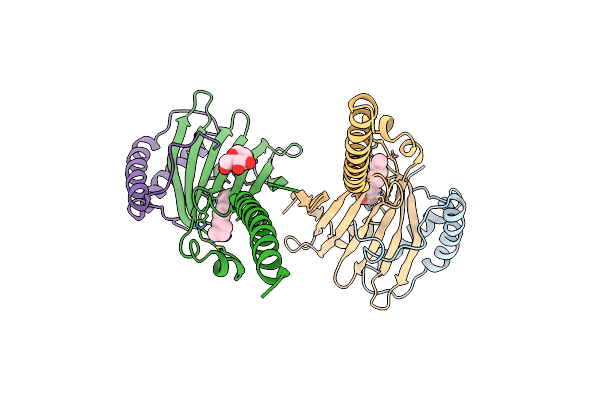 |
Crystal Structure Of The Human Mitochondrial Prelid1K58V-Triap1 Complex With Ps
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2019-03-20 Classification: LIPID TRANSPORT Ligands: LMT, P5S |
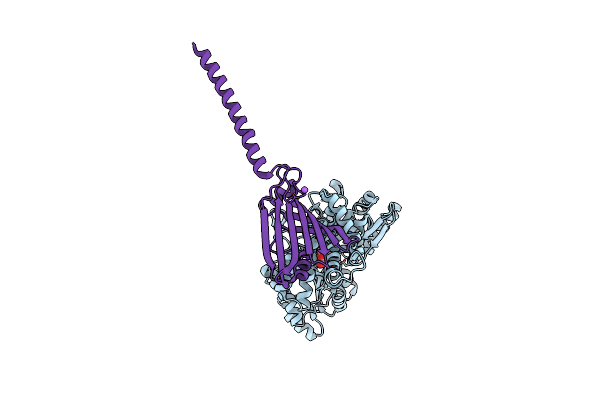 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2019-03-20 Classification: LIPID TRANSPORT |
 |
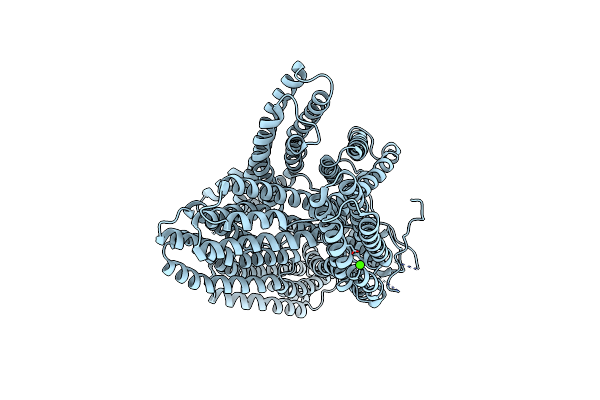
Structure Of Connexin-46 Intercellular Gap Junction Channel At 3.4 Angstrom Resolution By Cryoem
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2018-12-12 Classification: MEMBRANE PROTEIN |
 |
Structure Of Connexin-50 Intercellular Gap Junction Channel At 3.4 Angstrom Resolution By Cryoem
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2018-12-12 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Human Full-Length Vinculin-T12-A974K (Residues 1-1066)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-03-14 Classification: CELL ADHESION Ligands: CA |
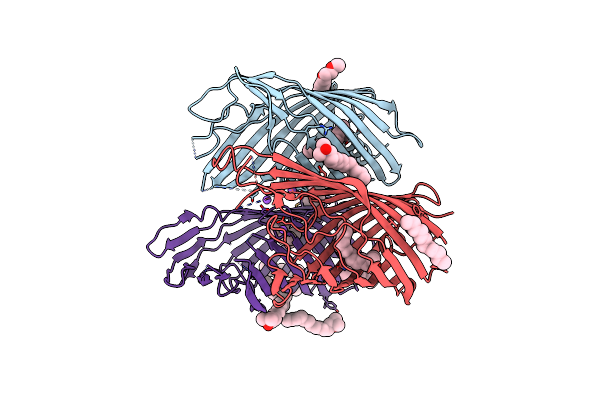 |
Crystal Structure Of The Pseudomonas Functional Amyloid Secretion Protein Fapf
Organism: Pseudomonas sp. uk4
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-08-23 Classification: MEMBRANE PROTEIN Ligands: LDA, C8E, NA |
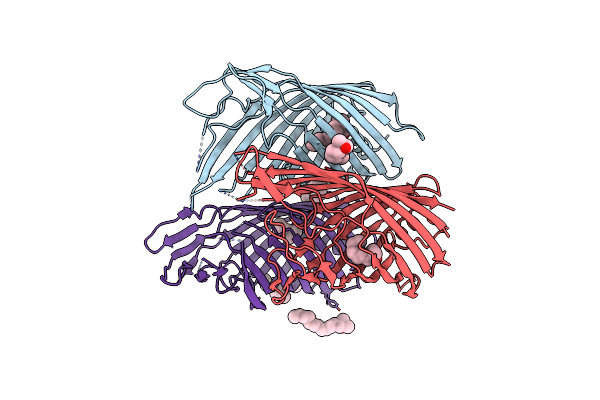 |
Organism: Pseudomonas sp. uk4
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2017-08-23 Classification: MEMBRANE PROTEIN Ligands: LDA, C8E |
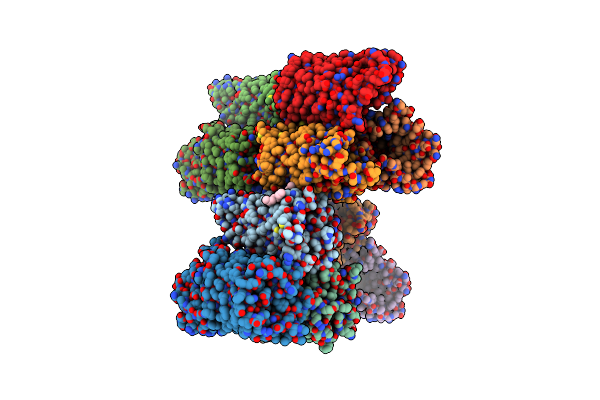 |
Crystal Structure Of The Pseudomonas Functional Amyloid Secretion Protein Fapf - R157A Mutant
Organism: Pseudomonas sp. uk4
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2017-08-23 Classification: MEMBRANE PROTEIN Ligands: C8E, LDA |
 |
Crystal Structure Of The Suscd Complex Bt2261-2264 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-12-21 Classification: MEMBRANE PROTEIN Ligands: KR0, NA, CA |
 |
Crystal Structure Of The Suscd Complex Bt2261-2264 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-12-21 Classification: MEMBRANE PROTEIN Ligands: MG, NA |
 |
Crystal Structure Of The Suscd Complex Bt2261-2264 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Release Date: 2016-12-21 Classification: MEMBRANE PROTEIN Ligands: MG, KR0, CA, C8E |
 |
Crystal Structure Of The Lipoprotein Bt2262 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-12-14 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Lipoprotein Bt2263 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-12-14 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: SO4, NA |

