Search Count: 13
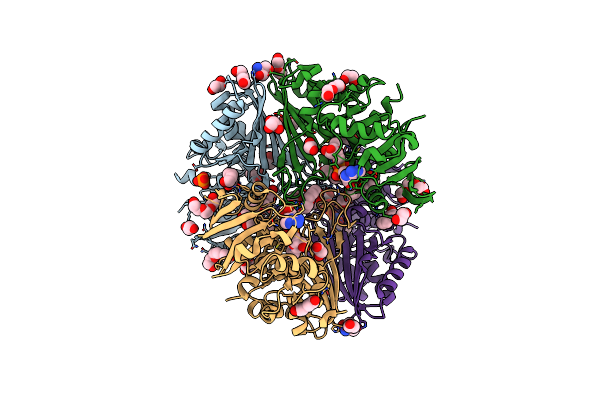 |
Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Nicotinamide Adenine Dinucleotide In The Presence Of Poly(Ethylene Glycol) At 2.20 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: NAD, PEG, PG4, EDO, PO4, PGE, TRS, 1PE, GOL |
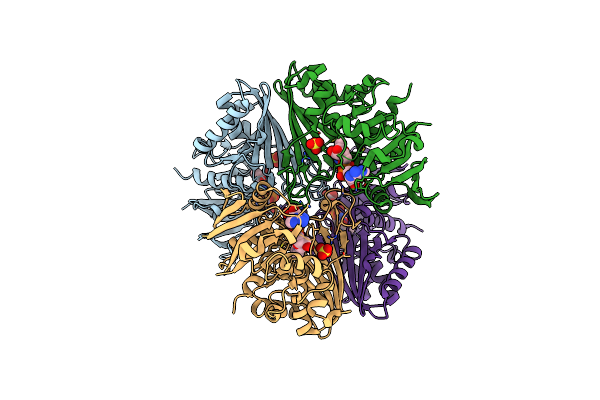 |
Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Nicotinamide Adenine Dinucleotide At 2.74 A Resolution.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
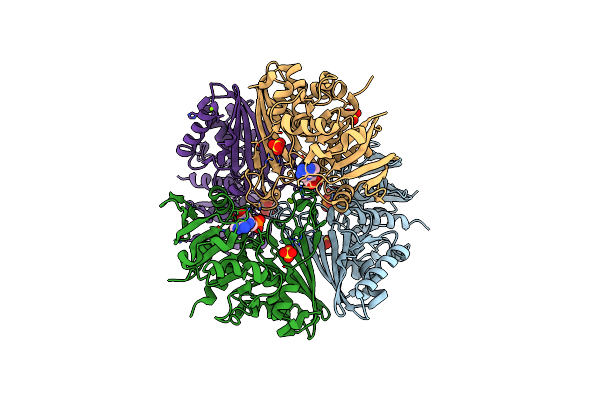 |
Crystal Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Adenosine Phosphate At 2.40 A Resolution.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: AMP, SO4, MG |
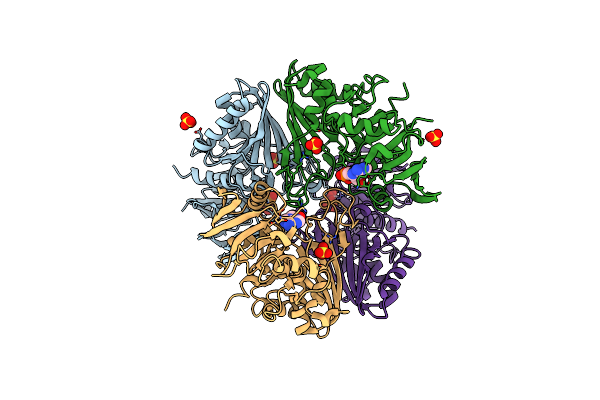 |
Crystal Structure Of The Complex Of Glyceraldehyde-3-Phosphate Dehydrogenase Of Type B From Acinetobacter Baumannii With Adenosine Monophosphate At 3.20 A Resolution.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-06-12 Classification: OXIDOREDUCTASE Ligands: AMP, SO4 |
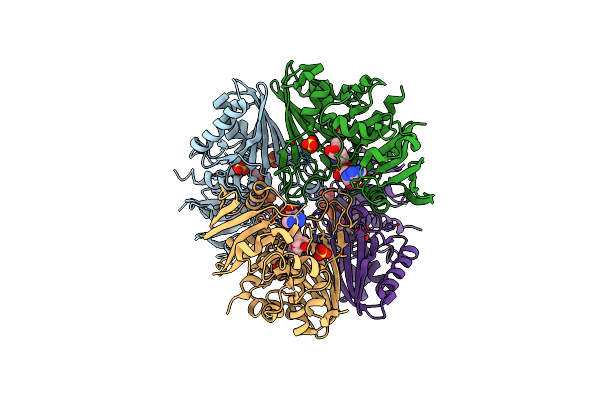 |
Structure Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii At 3.00 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-06-05 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
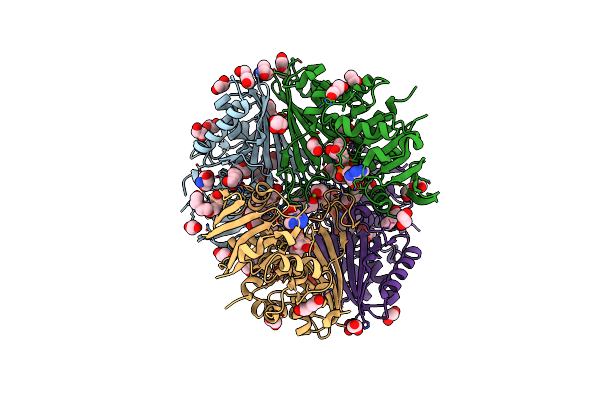 |
Crystal Structure Of Poly(Ethylene Glycol) Stabilized Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii At 2.30 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-05 Classification: OXIDOREDUCTASE Ligands: NAD, PEG, PG4, EDO, TRS, GOL, XPE, PGE, SO4, MG |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-01-17 Classification: HYDROLASE |
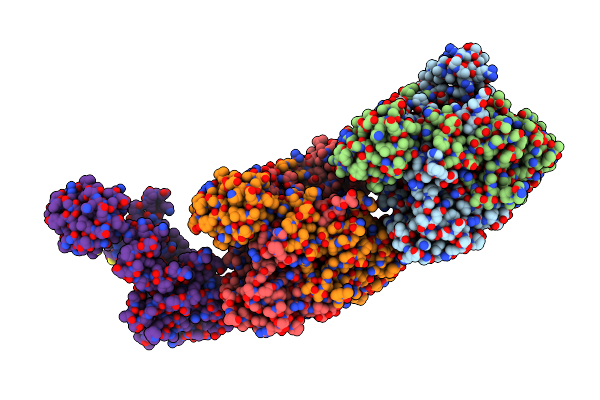 |
Crystal Structure Of Hypothetical Protein (Rv3272) From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2018-11-07 Classification: TRANSFERASE |
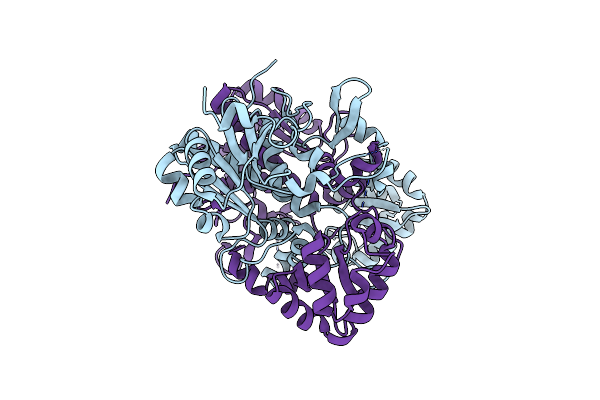 |
Crystal Structure Of D175A Mutant Of Rv3272 From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-11-07 Classification: TRANSFERASE Ligands: CL |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-11-07 Classification: TRANSFERASE Ligands: BEN, GOL |
 |
Crystal Structure Of The Ternary Complex Of Agrobacterium Radiobacter K84 Agnb2 Leurs-Trna-Leuams
Organism: Agrobacterium radiobacter k84, Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-02-17 Classification: LIGASE/RNA Ligands: ZN, LSS, SO4, MN |
 |
Ternary Complex Of E. Coli Leucyl-Trna Synthetase, Trna(Leu) And Toxic Moiety From Agrocin 84 (Tm84) In Aminoacylation-Like Conformation
Organism: Escherichia coli, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-01-30 Classification: LIGASE/RNA Ligands: ZN, 84T, MG |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2007-03-06 Classification: LYASE |

