Search Count: 25
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I5X |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I53 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I54 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I55 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: A1I5Y |
 |
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-09-04 Classification: VIRAL PROTEIN Ligands: K4J |
 |
Organism: Hepacivirus c
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-09-04 Classification: VIRAL PROTEIN Ligands: SO4, K4P |
 |
Organism: Hepatitis c virus genotype 1b (isolate bk)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-09-04 Classification: VIRAL PROTEIN Ligands: K4S |
 |
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2019-09-04 Classification: VIRAL PROTEIN Ligands: K4M |
 |
Cs-Rosetta Determined Structures Of The N-Terminal Domain Of Algf From P. Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: SOLUTION NMR Release Date: 2019-04-17 Classification: CELL ADHESION |
 |
Cs-Rosetta Determined Structures Of The C-Terminal Domain Of Algf From P. Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2019-04-17 Classification: CELL ADHESION |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-08-01 Classification: SIGNALING PROTEIN Ligands: ASP |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-08-01 Classification: SIGNALING PROTEIN Ligands: ASN |
 |
Organism: mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2018-07-25 Classification: SIGNALING PROTEIN Ligands: GLU |
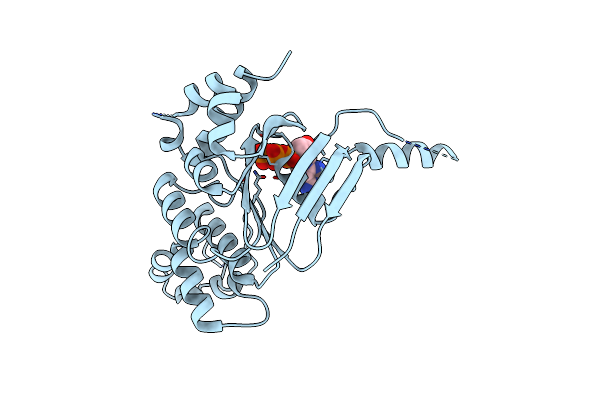 |
Human Cftr Aa389-678 (Nbd1), Deltaf508 With Three Solubilizing Mutations, Bound Atp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2015-11-11 Classification: ATP binding Protein Ligands: ATP, MG |
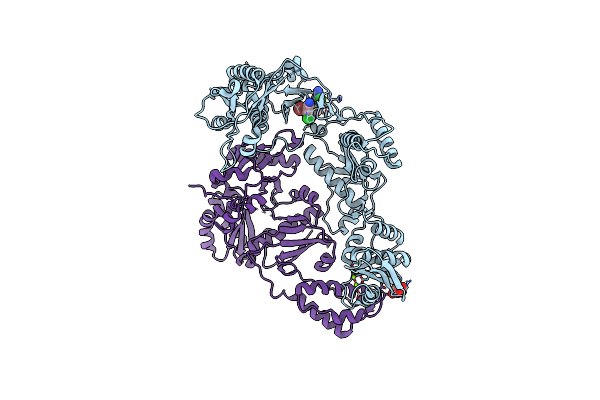 |
Organism: Hiv-1 m\:b_hxb2r
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2013-01-09 Classification: HYDROLASE Ligands: MG, TAR, WHU |
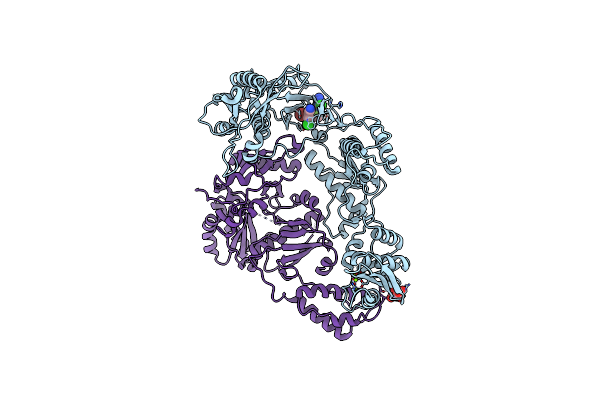 |
Organism: Hiv-1 m\:b_hxb2r
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2013-01-09 Classification: HYDROLASE Ligands: MG, SRT, WHU |
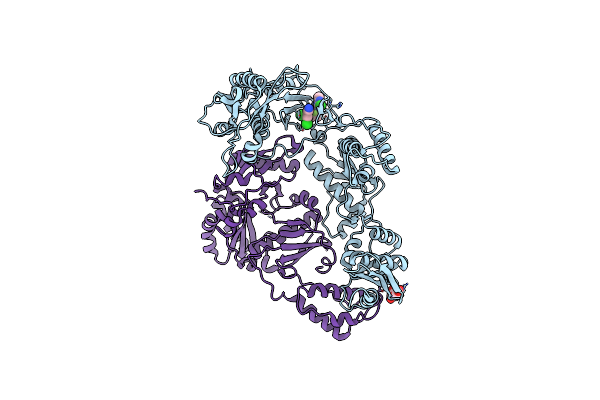 |
Organism: Hiv-1 m\:b_hxb2r
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-01-09 Classification: HYDROLASE Ligands: TAR, EUR |
 |
Organism: Hiv-1 m\:b_hxb2r
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2013-01-09 Classification: HYDROLASE Ligands: MG, TAR, CXD |
 |
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2011-12-21 Classification: STRUCTURAL PROTEIN |

