Search Count: 15
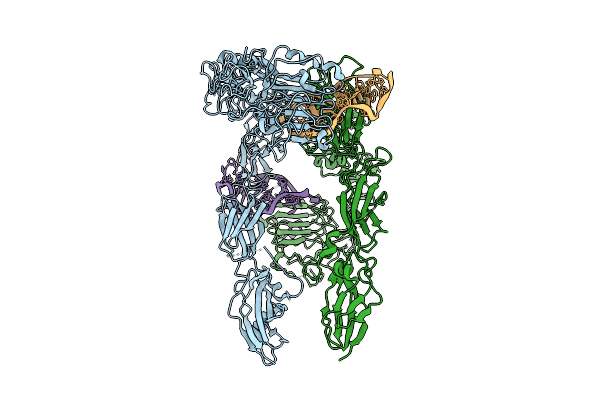 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
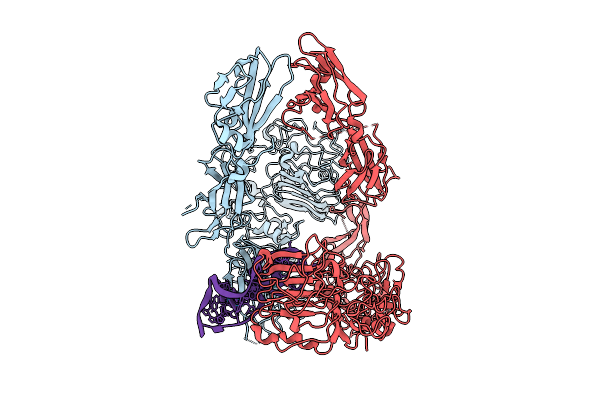 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
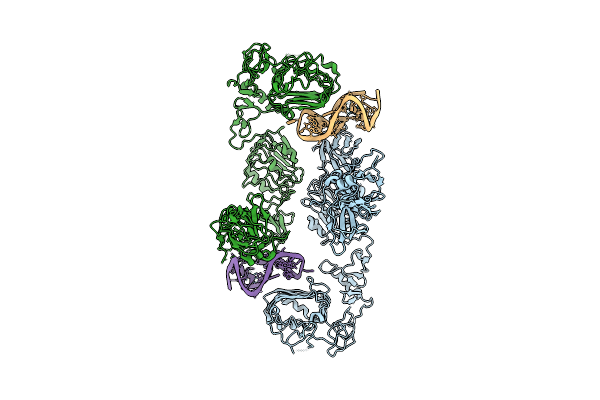 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
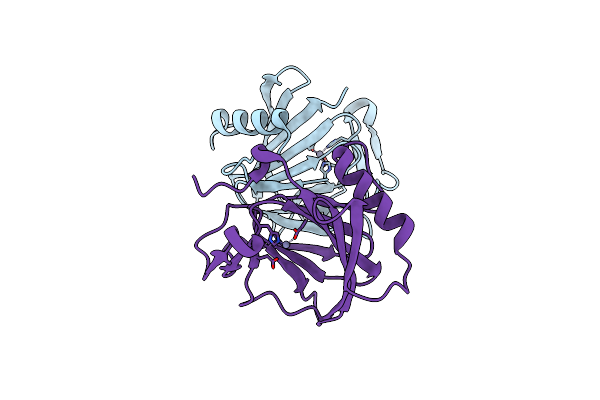 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-09-04 Classification: HYDROLASE |
 |
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-09-04 Classification: HYDROLASE |
 |
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2019-09-04 Classification: HYDROLASE |
 |
Crystal Structure Of Petase S121E, D186H, R280A Mutant From Ideonella Sakaiensis
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-09-04 Classification: HYDROLASE |
 |
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-02-14 Classification: HYDROLASE |
 |
Organism: Ideonella sakaiensis (strain 201-f6)
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2018-02-14 Classification: HYDROLASE |
 |
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-12-07 Classification: TRANSFERASE Ligands: GOL, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-06-22 Classification: HYDROLASE Ligands: PEG, PO4, HG |
 |
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-06-22 Classification: HYDROLASE Ligands: AYE, SO4, CL, DIO |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-03-18 Classification: SIGNALING PROTEIN Ligands: EDO, IOD, ACT, PG4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-03-18 Classification: SIGNALING PROTEIN Ligands: EDO |

