Search Count: 165
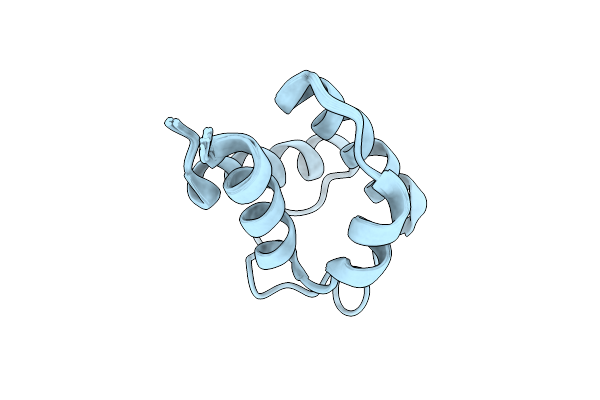 |
Solution Structure Of Holo Acyl Carrier Protein 2 (Apef) Of Aryl Polyene Biosynthesis From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: SOLUTION NMR Release Date: 2025-11-26 Classification: BIOSYNTHETIC PROTEIN |
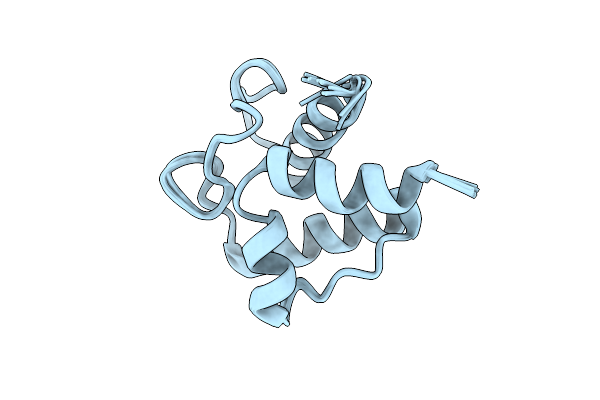 |
Solution Structure Of Holo Acyl Carrier Protein 1 (Apee) Of Aryl Polyene Biosynthesis From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: SOLUTION NMR Release Date: 2025-11-26 Classification: BIOSYNTHETIC PROTEIN |
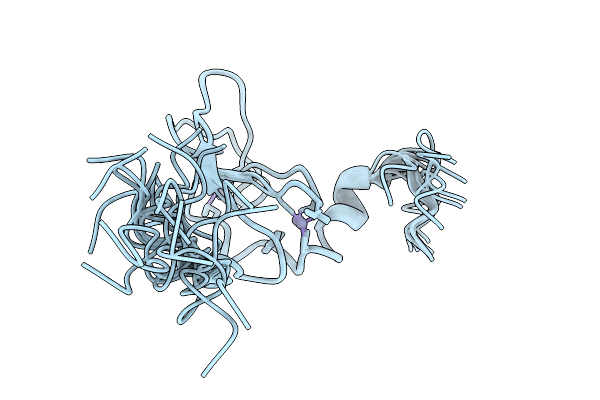 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: ZN |
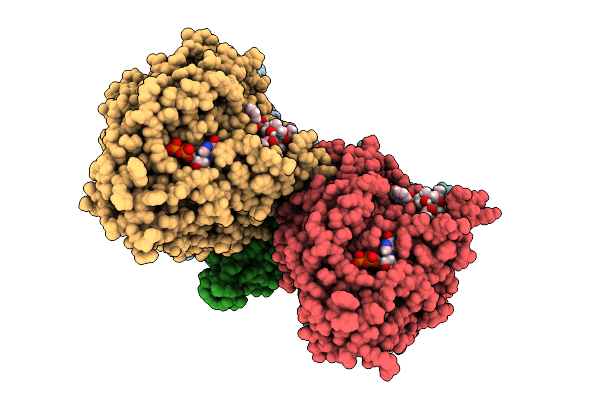 |
Cryo-Em Structure Of Taxol-Microtubules In Complex With The C1 Domain Of Gefh1
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: SIGNALING PROTEIN Ligands: ZN, GTP, MG, GDP, TA1 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
Organism: Klebsiella pneumoniae, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM Ligands: CIT, PLM, NAG |
 |
Organism: Klebsiella pneumoniae, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM Ligands: CIT, PLM, NAG |
 |
Cryo-Em Structure Of The G15C-R66C And T83C-T83C Diabody Complex (Cits-Diabody #7-Tlr3)
Organism: Klebsiella pneumoniae, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM Ligands: CIT, PLM, NAG |
 |
Cryo-Em Structure Of The Klebsiella Pneumoniae Cits (Citrate-Bound Occluded State)
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: CIT, PLM |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: PTY, NAG |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: PTY, NAG |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
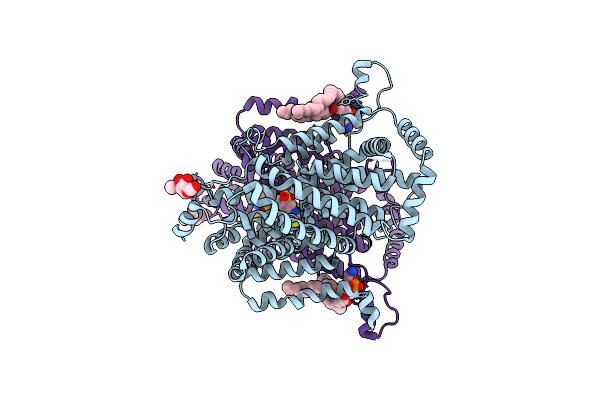
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: 4DS, PTY, NAG |
 |
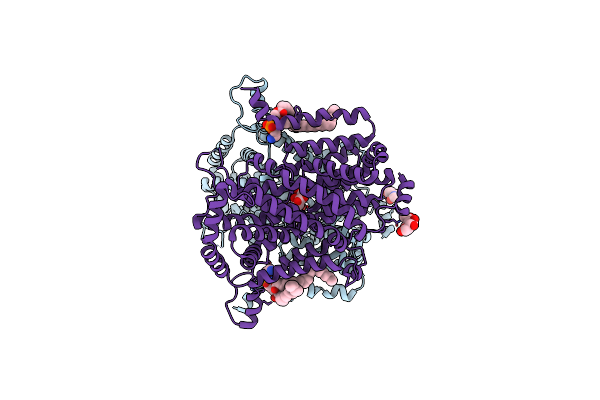
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: PTY, NAG, 4DS |
 |
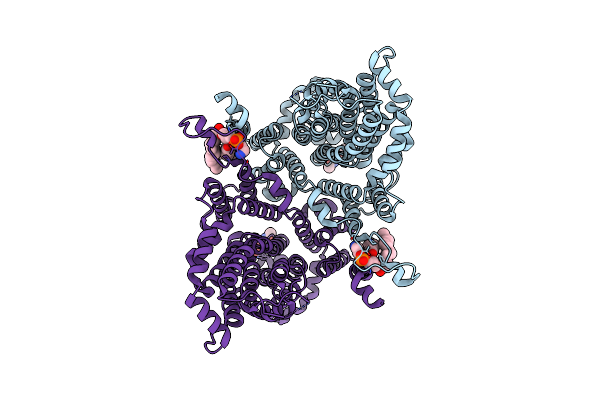
Cryo-Em Structure Of The Drosophila Indy (Citrate-Bound Inward-Occluded, Ph 6)
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: FLC, NAG, PTY |
 |
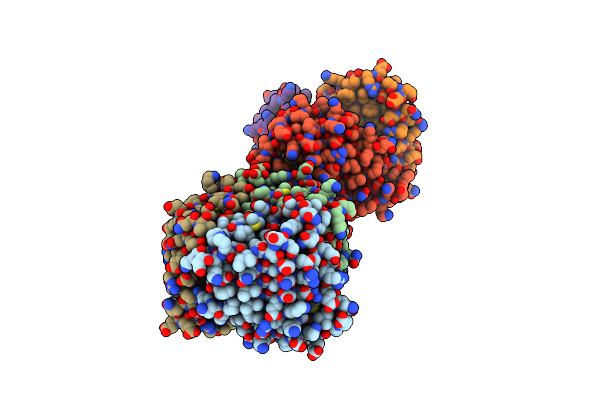
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: MEMBRANE PROTEIN Ligands: PTY, NAG |
 |
Crystal Structure Of The Gamma-Carbonic Anhydrase From The Polyextremophilic Bacterium Aeribacillus Pallidus
Organism: Aeribacillus pallidus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-11 Classification: METAL BINDING PROTEIN Ligands: ZN, SO4 |

