Search Count: 241
 |
Crystal Structure Of Glutamyl-Trna Synthetase Glurs From Pseudomonas Aeruginosa (Zinc Bound)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-12-27 Classification: LIGASE Ligands: FLC, SO4, ZN, 2PE, GOL |
 |
Solution Nmr Structure Of 9-Residue Rosetta-Designed Cyclic Peptide D9.16 In D6-Dmso With Cis/Trans Switching
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.31 In D6-Dmso With Cis/Trans Switching (A-Cc Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In D6-Dmso With Cis/Trans Switching
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In 50% D6-Dmso And 50% Water With Cis/Trans Switching (Cc Conformation, 50%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 9-Residue Rosetta-Designed Cyclic Peptide D9.16 In Cdcl3 With Cis/Trans Switching (A-Tt Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.31 In Cdcl3 With Cis/Trans Switching
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In Cdcl3 With Cis/Trans Switching (Tt Conformation, 47%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2022-09-14 Classification: DE NOVO PROTEIN Ligands: HOH |
 |
Solution Nmr Structure Of 9-Residue Rosetta-Designed Cyclic Peptide D9.16 In Cdcl3 With Cis/Trans Switching (B-Tc Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.31 In D6-Dmso With Cis/Trans Switching (B-Ct Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In 50% D6-Dmso And 50% Water With Cis/Trans Switching (Cc Conformation, 50%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In Cdcl3 With Cis/Trans Switching (Tc Conformation, 53%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
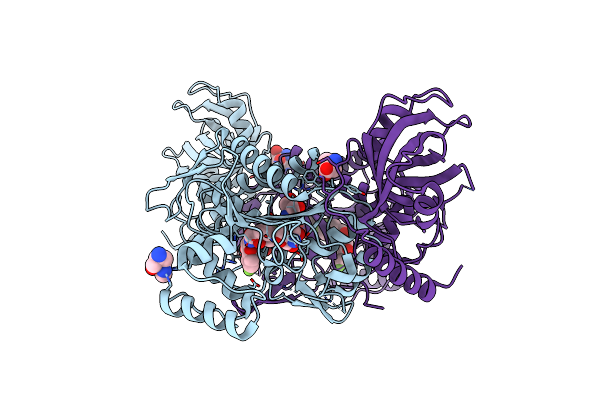
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) From Chlamydia Trachomatis With Bound Nad
Organism: Chlamydia trachomatis (strain d/uw-3/cx)
Method: X-RAY DIFFRACTION Release Date: 2019-04-24 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, PO4, PEG, CL |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Plasmodium Falciparum Bound To A Difluoro Cyclohexyl Chromone Ligand
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: FYB, LYS, PEG, HIS, TRS |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Plasmodium Falciparum Complexed With A Chromone Ligand
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: FYE |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Cryptosporidium Parvum Complexed With L-Lysine And A Difluoro Cyclohexyl Chromone Ligand
Organism: Cryptosporidium parvum (strain iowa ii)
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: LYS, FYB, TRS |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-03-13 Classification: LIGASE/INHIBITOR Ligands: 9X0, CO, FMT, MLA, LYS |
 |
Calcium-Dependent Protein Kinase 1 From Toxoplasma Gondii (Tgcdpk1) In Complex With Inhibitor Uw1553
Organism: Toxoplasma gondii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-12-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: UW5 |
 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae Bound To Nad
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-05-10 Classification: OXIDOREDUCTASE Ligands: NAD, CL |

