Search Count: 23
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: HYDROLASE Ligands: GDP, ANP, MG, GNP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: HYDROLASE Ligands: GDP, ANP, MG, GNP |
 |
Crystal Structure Of Germination Protease From The Spore-Forming Bacterium Paenisporosarcina Sp. Tg-20 In Its Inactive Form
Organism: Paenisporosarcina sp.
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-03-31 Classification: PROTEASE |
 |
Organism: Vibrio sp. (strain ejy3)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-09-27 Classification: ISOMERASE Ligands: MG, ACY |
 |
Organism: Vibrio sp. (strain ejy3)
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2017-09-27 Classification: ISOMERASE Ligands: MG |
 |
Organism: Vibrio sp. (strain ejy3)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-09-27 Classification: ISOMERASE Ligands: MG |
 |
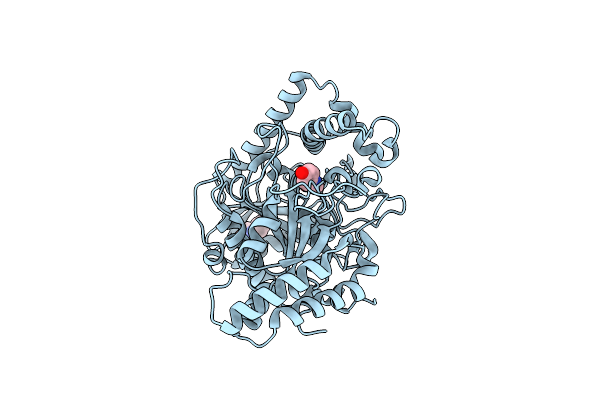
The Crystal Structure Of A Bacterial Aryl Acylamidase Belonging To The Amidase Signature (As) Enzymes Family
Organism: Bacterium csbl00001
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: PO4 |
 |
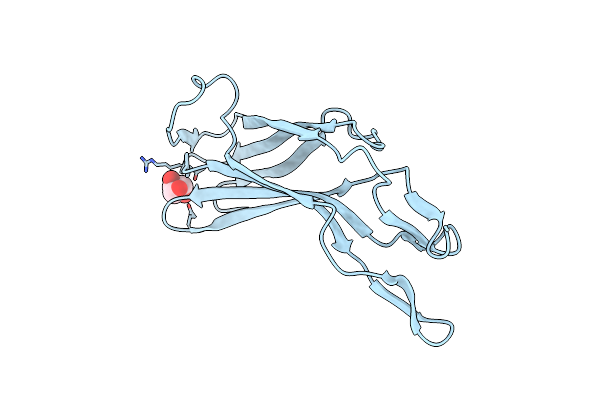
The Crystal Structure Of A Bacterial Aryl Acylamidase Belonging To The Amidase Signature (As) Enzymes Family
Organism: Bacterium csbl00001
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: CXS, TYL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-02-05 Classification: PROTEIN BINDING Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-02-05 Classification: ANTITUMOR PROTEIN/PROTEIN BINDING |
 |
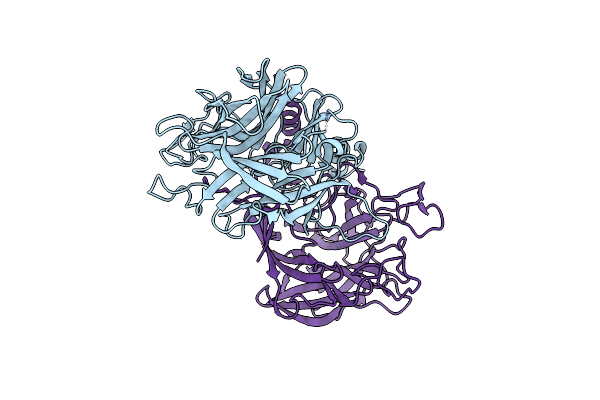
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-02-05 Classification: ANTITUMOR PROTEIN/PROTEIN BINDING |
 |
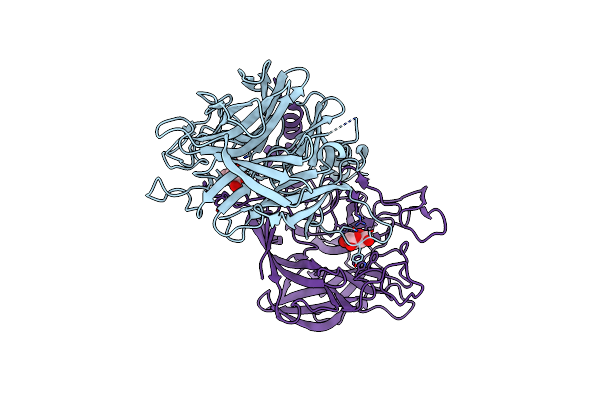
Crystal Structure Of Alpha-Neoagarobiose Hydrolase (Alpha-Nabh) From Saccharophagus Degradans 2-40
Organism: Saccharophagus degradans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-02-01 Classification: HYDROLASE |
 |
Crystal Structure Of Alpha-Neoagarobiose Hydrolase (Alpha-Nabh) In Complex With Alpha-D-Galactopyranose From Saccharophagus Degradans 2-40
Organism: Saccharophagus degradans
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-02-01 Classification: HYDROLASE Ligands: GLA |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: MES |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
Structure Of A Deazaflavin-Dependent Nitroreductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42 |
 |
Structure Of A Deazaflavin-Dependent Reductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42, SO4 |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-12-07 Classification: HYDROLASE |

