Search Count: 27
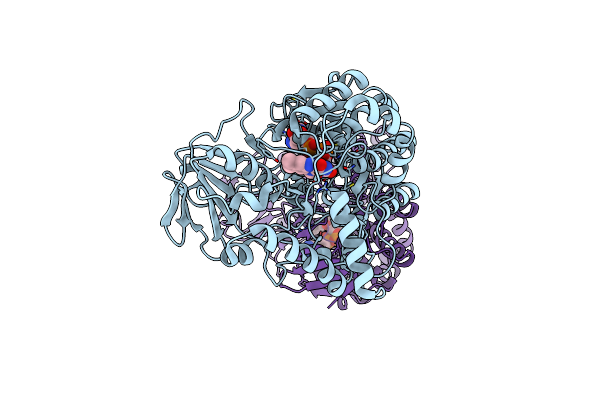 |
Crystal Structure Of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans
Organism: Parvibaculum lavamentivorans (strain ds-1 / dsm 13023 / ncimb 13966)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-04-10 Classification: OXIDOREDUCTASE Ligands: FAD, MG |
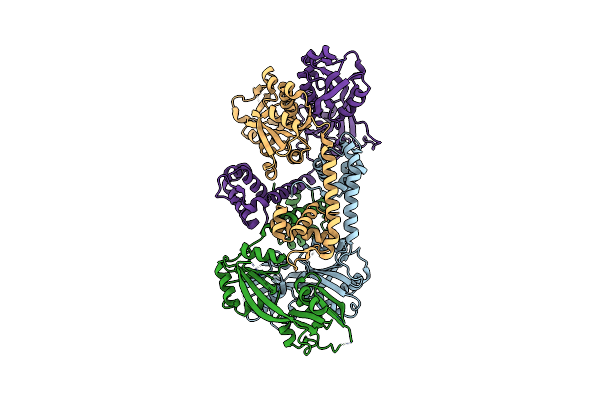 |
Organism: Vibrio vulnificus cmcp6
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-03-28 Classification: TRANSCRIPTION Ligands: BR |
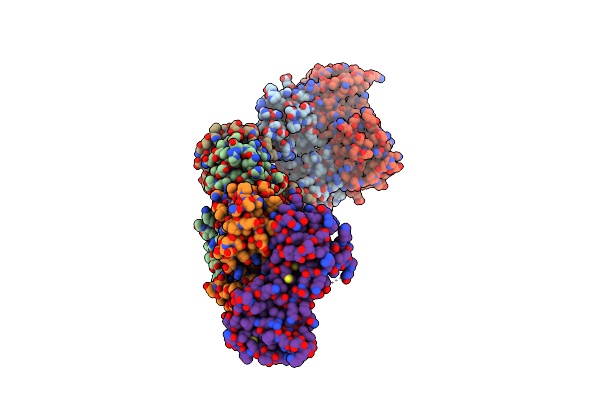 |
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2017-05-10 Classification: TRANSCRIPTION |
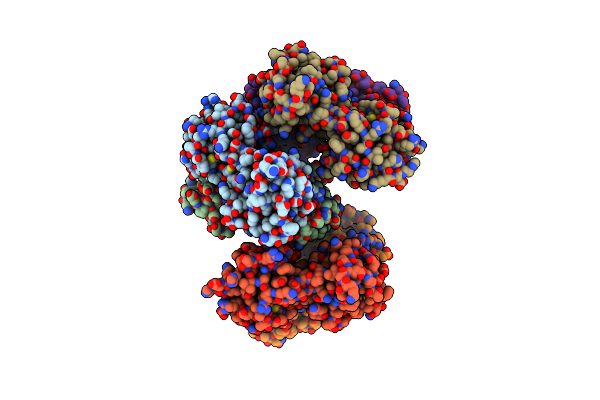 |
Regulatory Domain Of Aphb Treated With Cumene Hydroperoxide From Vibrio Vulnificus
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-05-10 Classification: TRANSCRIPTION |
 |
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2017-01-11 Classification: TRANSCRIPTION |
 |
T. Brucei Farnesyl Diphosphate Synthase Complexed With Bisphosphonate Bph-597
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-10-28 Classification: TRANSFERASE Ligands: QAF, MG |
 |
T. Brucei Farnesyl Diphosphate Synthase Complexed With Bisphosphonate Bph-1238
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2015-10-28 Classification: TRANSFERASE Ligands: MG, PVZ |
 |
T. Brucei Farnesyl Diphosphate Synthase Complexed With Bisphosphonate Bph-1326
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2015-10-28 Classification: TRANSFERASE Ligands: G76, MG |
 |
Crystal Structure Of Ketosteroid Isomerase Containing Y32F, D40N, Y57F And Y119F Mutations In The Equilenin-Bound Form
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-05-20 Classification: ISOMERASE Ligands: EQU |
 |
Crystal Structure Of A Ras-Like Protein From Cryphonectria Parasitica In Complex With Gdp
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-06-05 Classification: SIGNALING PROTEIN Ligands: GDP, MG |
 |
Crystal Structure Of Hy5 Leucine Zipper Homodimer From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-03-20 Classification: TRANSCRIPTION |
 |
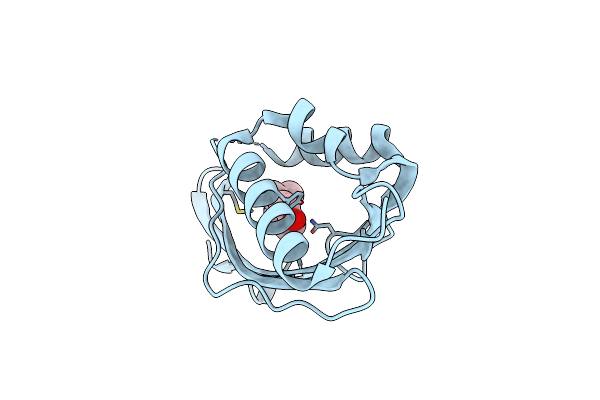
Crystal Structure Of D103E Mutant With Equilenineof Ksi In Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-06-17 Classification: ISOMERASE Ligands: EQU |
 |
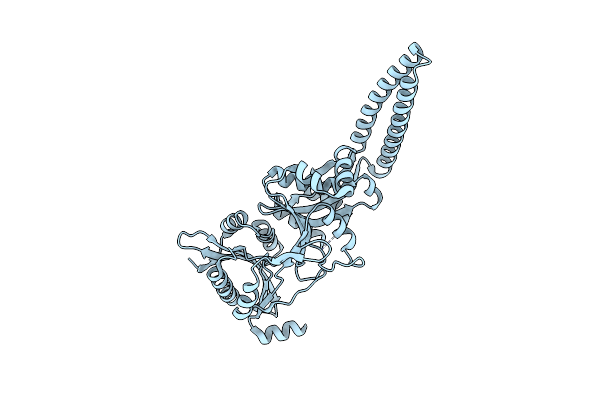
High Resolution Crystal Structure Of Pi Delta-5-3-Ketosteroid Isomerase Mutants Y30F/Y55F/Y115F/D38N (Y32F/Y57F/Y119F/D40N, Pi Numbering)Complexed With Equilenin At 2.1 A Resolution
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-01-23 Classification: ISOMERASE Ligands: EQU |
 |
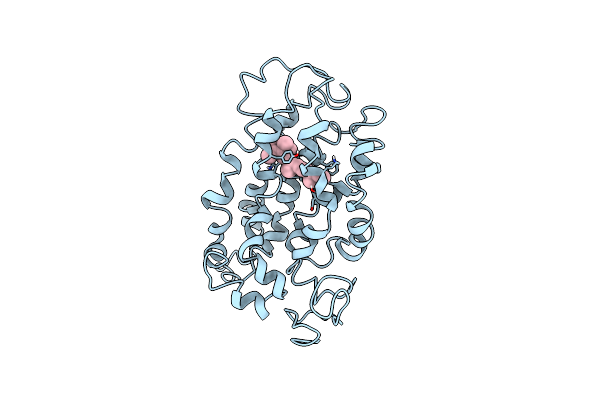
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-09-04 Classification: ANTIBIOTIC INHIBITOR |
 |
 |
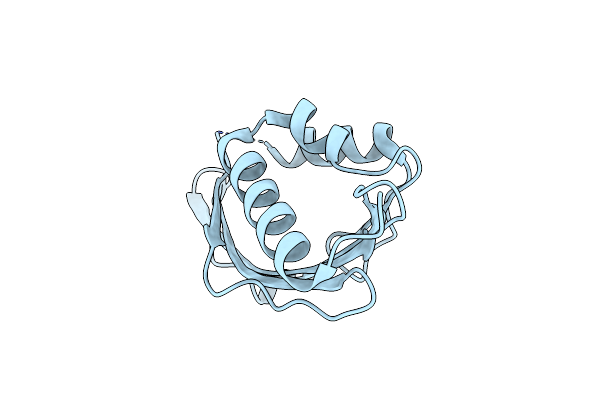
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-06-12 Classification: HYDROLASE Ligands: ZN |
 |
Pseudoreversion Of The Catalytic Activity Of Y14F By The Additional Tyrosine-To-Phenylalanine Substitution(S) In The Hydrogen Bond Network Of Delta-5-3-Ketosteroid Isomerase From Pheudomonas Putida Biotype B
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-11-01 Classification: ISOMERASE |
 |
Crystal Structure Of Ketosteroid Isomerase From Pseudomonas Putida ; Triple Mutant Y16F/Y32F/Y57F
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-10-05 Classification: ISOMERASE |
 |
Organism: Bos taurus
Method: SOLUTION NMR Release Date: 2001-10-05 Classification: SIGNALING PROTEIN, MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2001-03-14 Classification: OXIDOREDUCTASE Ligands: FAD |

