Search Count: 45
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2023-01-04 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-01-04 Classification: ELECTRON TRANSPORT Ligands: TRS, CU |
 |
Organism: Blastobotrys adeninivorans
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-17 Classification: LYASE Ligands: 4NC, K, CO |
 |
Crystal Structure Of The Pea Pathogenicity Protein 2 From Madurella Mycetomatis
Organism: Madurella mycetomatis
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-02-17 Classification: LYASE Ligands: K, CA |
 |
Crystal Structure Of The Pea Pathogenicity Protein 2 From Madurella Mycetomatis Complexed With 4-Nitrocatechol
Organism: Madurella mycetomatis
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-02-17 Classification: LYASE Ligands: 4NC, K, CA |
 |
Organism: Blastobotrys adeninivorans
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-02-17 Classification: LYASE Ligands: K |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-05-23 Classification: TRANSFERASE Ligands: MN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2018-05-23 Classification: TRANSFERASE Ligands: MN, UDP, TRS, NO3 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2018-05-23 Classification: TRANSFERASE Ligands: MN, UDP, GOL, TRS, NO3 |
 |
Crystal Structure Of Udp-Glucosyltransferase, Ugt74F2, With Udp And Salicylic Acid
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2017-05-03 Classification: TRANSFERASE Ligands: BGC, UDP, SAL |
 |
Crystal Structure Of Udp-Glucosyltransferase, Ugt74F2 (T15S), With Udp And Salicylic Acid
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-05-03 Classification: TRANSFERASE Ligands: GLN, BGC, UDP, SAL |
 |
Crystal Structure Of Udp-Glucosyltransferase, Ugt74F2, With Udp And 2-Bromobenzoic Acid
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-05-03 Classification: TRANSFERASE Ligands: BGC, UDP, 7WV |
 |
Crystal Structure Of Udp-Glucosyltransferase, Ugt74F2 (T15S), With Udp And 2-Bromobenzoic Acid
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-05-03 Classification: TRANSFERASE Ligands: BGC, UDP, 7WV |
 |
Crystal Structure Of Udp-Glucosyltransferase, Ugt74F2 (T15A), With Udp And 2-Bromobenzoic Acid
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-05-03 Classification: TRANSFERASE Ligands: BGC, UDP, 7WV |
 |
Organism: Aequorea victoria, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-08-17 Classification: APOPTOSIS |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: SO4, NA |
 |
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: MG |
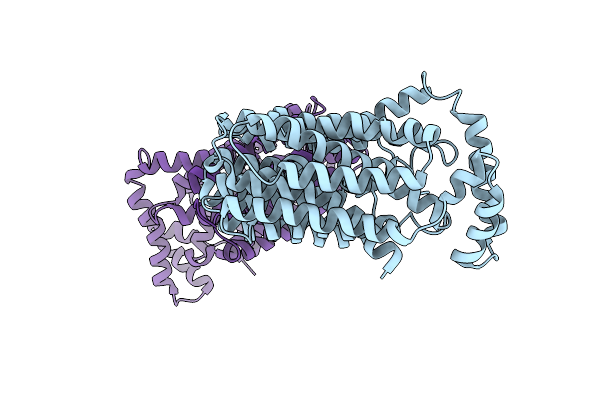 |
The Inward-Facing Structure Of The Glucose Transporter From Staphylococcus Epidermidis
Organism: Staphylococcus epidermidis (strain atcc 12228 / fda pci 1200)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2013-10-16 Classification: transport protein, membrane protein |
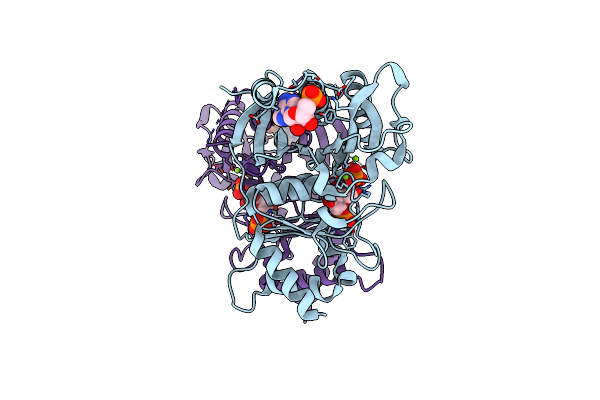 |
Crystal Structure Of Amp Complexes Of Porcine Liver Fructose-1,6-Bisphosphatase Mutant I10D In T-State
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-07-24 Classification: HYDROLASE Ligands: F6P, PO4, MG, AMP |
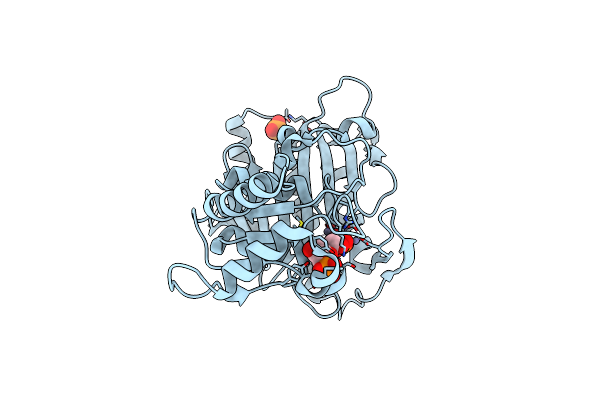 |
Mechanism Of Displacement Of A Catalytically Essential Loop From The Active Site Of Fructose-1,6-Bisphosphatase
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: F6P, PO4, ZN |

