Search Count: 219
 |
Organism: Clostridium phage phicd111
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ANTIBIOTIC Ligands: ZN |
 |
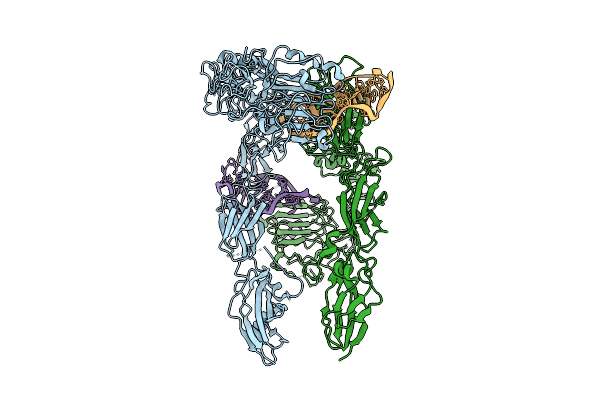
Cryo-Em Structure Of Outward State Anhydromuropeptide Permease (Ampg) Complex With Glcnac-1,6-Anhmurnac
Organism: Homo sapiens, Synthetic construct, Yokenella regensburgei, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: 2YP |
 |
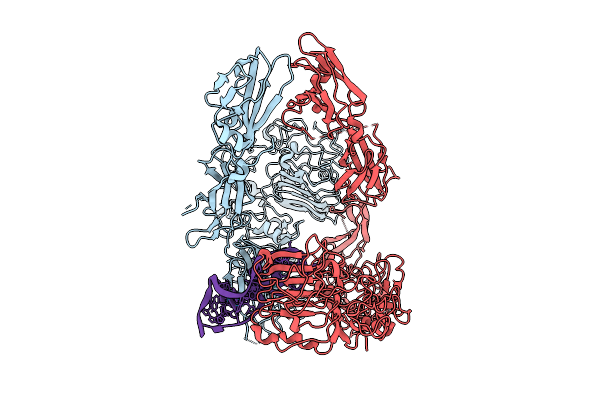
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
 |
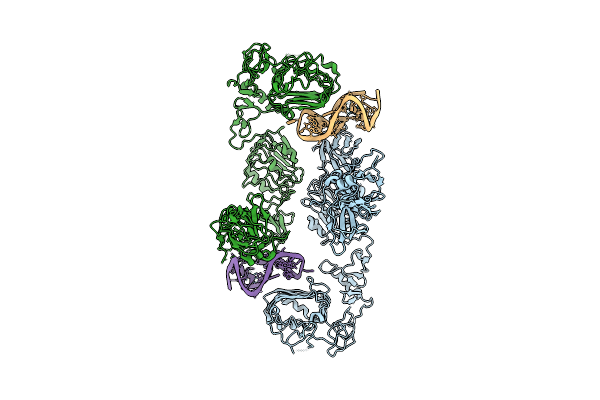
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Yokenella regensburgei
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Inward-Facing Anhydromuropeptide Permease (Ampg) In Complex With Glcnac-1,6-Anhmurnac
Organism: Yokenella regensburgei
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: 2YP |
 |
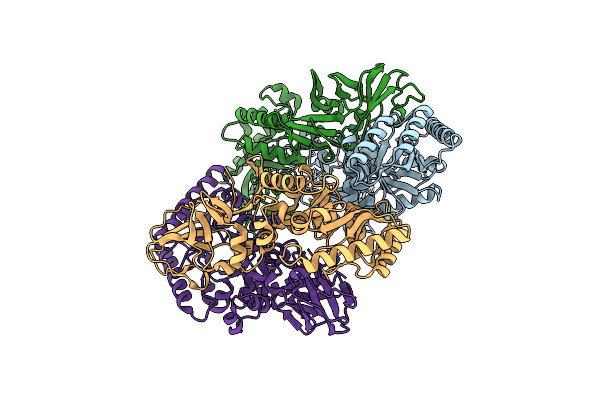
Cryo-Em Structure Of Outward State Anhydromuropeptide Permease (Ampg) G50W/L269W
Organism: Yokenella regensburgei, Escherichia coli, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: PEPTIDE BINDING PROTEIN |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, ALA, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
De Novo Designed Bbf-14 Beta Barrel With Computationally Designed Bbf-14_B4 Binder
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: DE NOVO PROTEIN Ligands: P6G, K, PG4, CL |
 |
Der F 21 Dust Mite Allergen With Computationally Designed Derf21_B10 Binder
Organism: Dermatophagoides farinae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: DE NOVO PROTEIN |
 |
Organism: Dermatophagoides farinae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: DE NOVO PROTEIN |
 |
Organism: Dermatophagoides farinae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: DE NOVO PROTEIN |
 |
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: CLR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2024-06-19 Classification: HYDROLASE/RNA Ligands: PO4, CA, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-05-22 Classification: HYDROLASE/RNA Ligands: GOL, PO4, PEG |

