Search Count: 26
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: PO4, DMS, O5J |
 |
E.Coli Lpxa In Complex With Udp-3-O-(R-3-Hydroxymyristoyl)-Glcnac And Compound 2
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: U20, PO4, DMS, O5P |
 |
E.Coli Lpxa In Complex With Udp-3-O-(R-3-Hydroxymyristoyl)-Glcnac And Compound 6
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: U20, PO4, DMS, O5M |
 |
E.Coli Lpxa In Complex With Udp-3-O-(R-3-Hydroxymyristoyl)-Glcnac And Compound 7
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: U20, PO4, DMS, O5G |
 |
E.Coli Lpxa In Complex With Udp-3-O-(R-3-Hydroxymyristoyl)-Glcnac And Compound 8
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: U20, PO4, DMS, O5D |
 |
Crystal Structure Of Fgf Receptor 2 Tyrosine Kinase Domain Harboring The D650V Activating Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-02-22 Classification: TRANSFERASE Ligands: SO4, ANP |
 |
Crystal Structure Of The Tyrosine Kinase Domain Of Fgf Receptor 2 Harboring A E565A/D650V Double Gain-Of-Function Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-02-22 Classification: TRANSFERASE Ligands: SO4, ACP, MG |
 |
Crystal Structure Of The Tyrosine Kinase Domain Of Fgf Receptor 2 Harboring A N549H/E565A Double Gain-Of-Function Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2017-02-22 Classification: TRANSFERASE Ligands: SO4, ACP, MG |
 |
Crystal Structure Of The Tyrosine Kinase Domain Of Fgf Receptor 2 Harboring An E565A/K659M Double Gain-Of-Function Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-02-22 Classification: TRANSFERASE Ligands: FLC, SO4, ACP |
 |
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
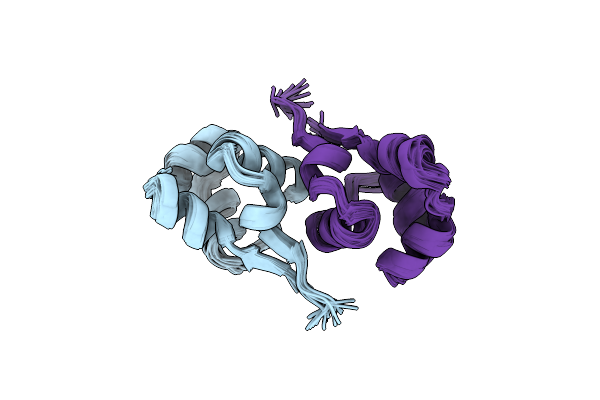 |
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
Noe-Based 3D Structure Of The Predissociated Homodimer Of Cylr2 In Equilibrium With Monomer At 266K (-7 Celsius Degrees)
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
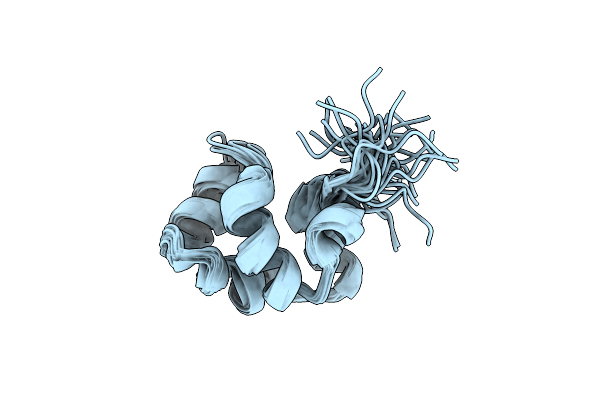
Noe-Based 3D Structure Of The Monomer Of Cylr2 In Equilibrium With Predissociated Homodimer At 266K (-7 Celsius Degrees)
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
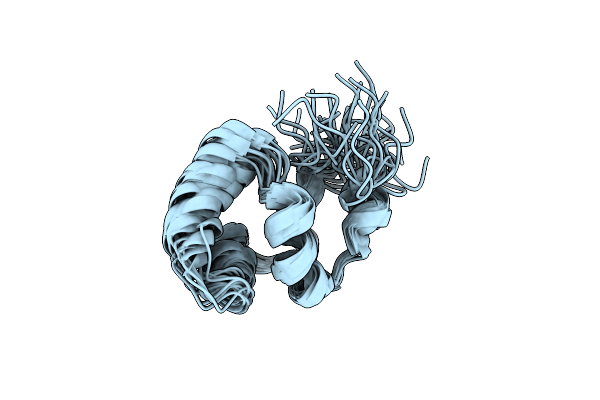
Noe-Based 3D Structure Of The Monomeric Intermediate Of Cylr2 At 262K (-11 Celsius Degrees)
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
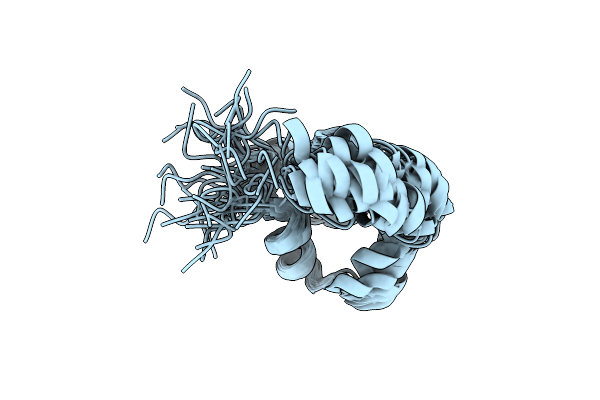
Noe-Based 3D Structure Of The Monomeric Partially-Folded Intermediate Of Cylr2 At 259K (-14 Celsius Degrees)
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
Noe-Based 3D Structure Of The Monomeric Partially-Folded Intermediate Of Cylr2 At 257K (-16 Celsius Degrees)
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2012-10-03 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-03 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-03 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2011-02-09 Classification: TRANSCRIPTION Ligands: GOL |

