Search Count: 25
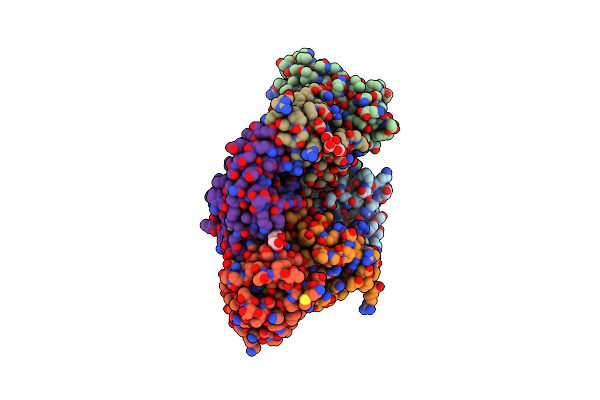 |
Crystal Structure Of Human Tigit(23-129) In Complex With The Scfv Fragment Of Anti-Tigit Antibody Mg1131
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-03-02 Classification: IMMUNE SYSTEM Ligands: FLC |
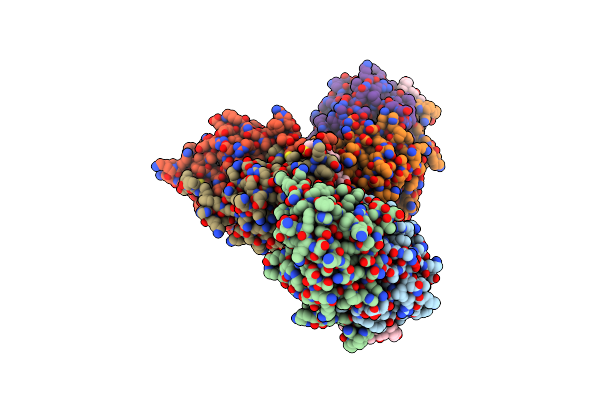 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-01-13 Classification: OXIDOREDUCTASE Ligands: GOL, CPS |
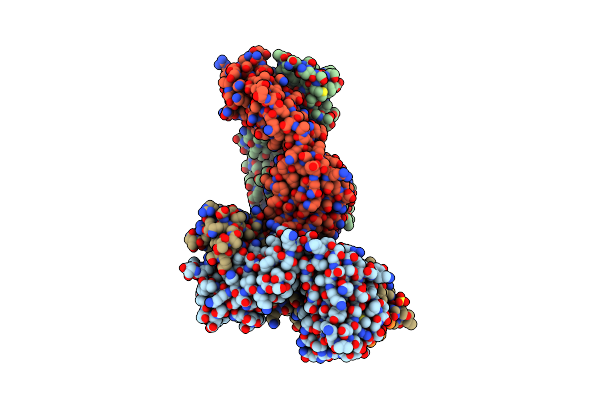 |
Organism: Influenza a virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-10-28 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Three-Dimensional Structure Of The Extracellular Domain Of Matrix Protein 2 Of Influenza A Virus
Organism: Influenza a virus (a/puerto rico/8/1934(h1n1)), Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-10-22 Classification: IMMUNE SYSTEM |
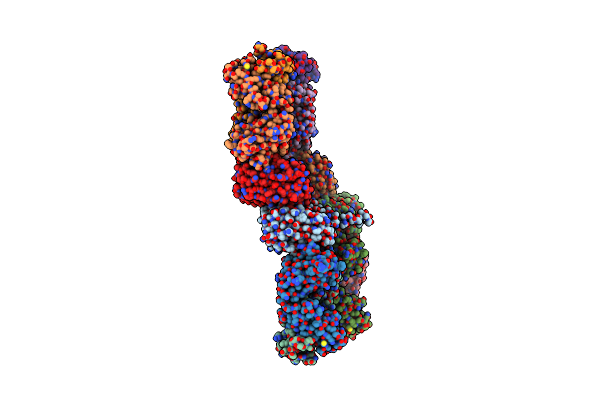 |
Insight Into Highly Conserved H1 Subtype-Specific Epitopes In Influenza Virus Hemagglutinin
Organism: Influenza a virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-04-09 Classification: IMMUNE SYSTEM Ligands: CA |
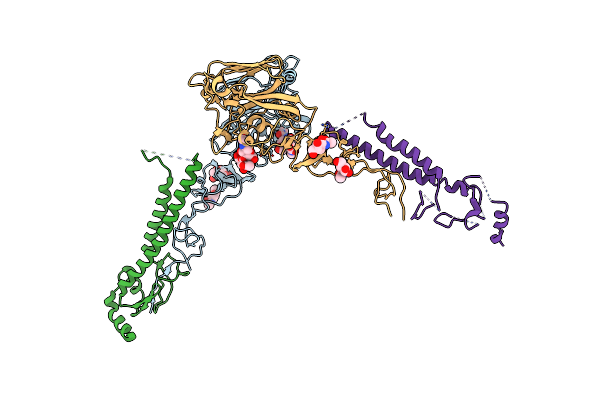 |
Structures Of Monomeric Hemagglutinin And Its Complex With An Fab Fragment Of A Neutralizing Antibody That Binds To H1 Subtype Influenza Viruses: Molecular Basis Of Infectivity Of 2009 Pandemic H1N1 Influenza A Viruses
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-05-22 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structures Of Monomeric Hemagglutinin And Its Complex With An Fab Fragment Of A Neutralizing Antibody That Binds To H1 Subtype Influenza Viruses: Molecular Basis Of Infectivity Of 2009 Pandemic H1N1 Influenza A Viruses
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-05-22 Classification: VIRAL PROTEIN |
 |
Organism: Influenza a virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2013-05-15 Classification: IMMUNE SYSTEM |
 |
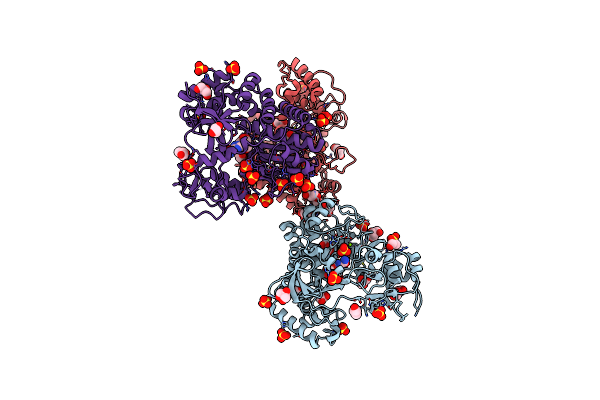
Crystal Structure Of Murine Norovirus Rna Dependent Rna Polymerase In Complex With 2Thiouridine(2Tu)
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2012-05-09 Classification: TRANSFERASE Ligands: 2TU, SO4, GOL, MG |
 |
Crystal Structure Of Murine Norovirus Rna Dependent Rna Polymerase In Complex With Ribavirin
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-05-09 Classification: TRANSFERASE Ligands: RBV, SO4, MG, GOL |
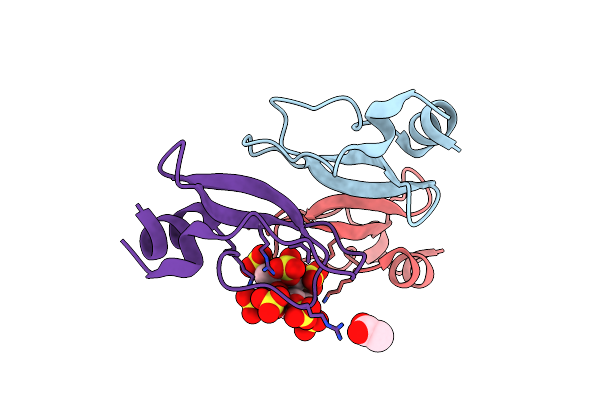 |
Crystal Structure Of Aprotinin In Complex With Sucrose Octasulfate: Unusual Interactions And Implication For Heparin Binding
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-09-15 Classification: HYDROLASE INHIBITOR Ligands: ACT |
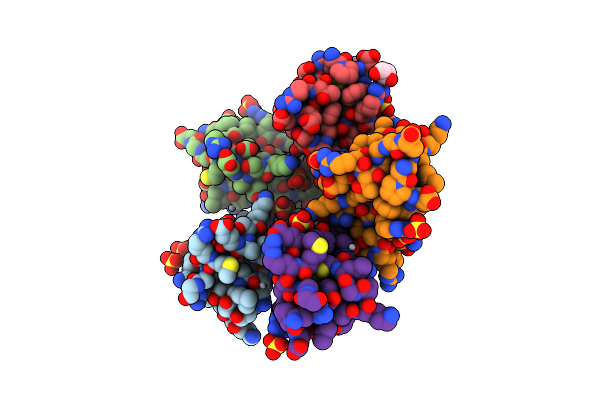 |
Crystal Structure Of Aprotinin In Complex With Sucrose Octasulfate: Unusual Interactions And Implication For Heparin Binding
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-06-23 Classification: HYDROLASE INHIBITOR Ligands: SO4, HG, GOL |
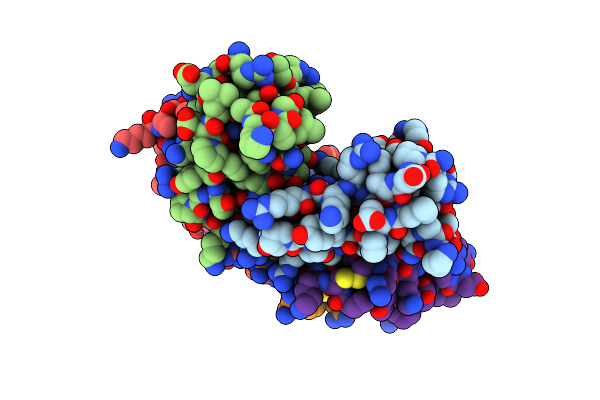 |
Crystal Structure Of Aprotinin In Complex With Sucrose Octasulfate: Unusual Interactions And Implication For Heparin Binding
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-06-23 Classification: HYDROLASE INHIBITOR |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-01-26 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-01-26 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-07-28 Classification: OXIDOREDUCTASE Ligands: FE, GOL |
 |
Structural Basis For The Iron Uptake Mechanism Of Helicobacter Pylori Ferritin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
Structural Basis For The Iron Uptake Mechanism Of Helicobacter Pylori Ferritin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: FE, IPA, GOL |
 |
Structural Basis For The Iron Uptake Mechanism Of Helicobacter Pylori Ferritin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: FE, GOL |
 |
Structural Basis For The Iron Uptake Mechanism Of Helicobacter Pylori Ferritin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: FE, GOL |

