Search Count: 20
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Pseudomonas Fluorescens Kmo In Complex With Ro 61-8048
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: FAD, 7ZR |
 |
Crystal Structure Of Saccharomyces Cerevisiae Kmo In Complex With Ro 61-8048
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: FAD, 7ZR |
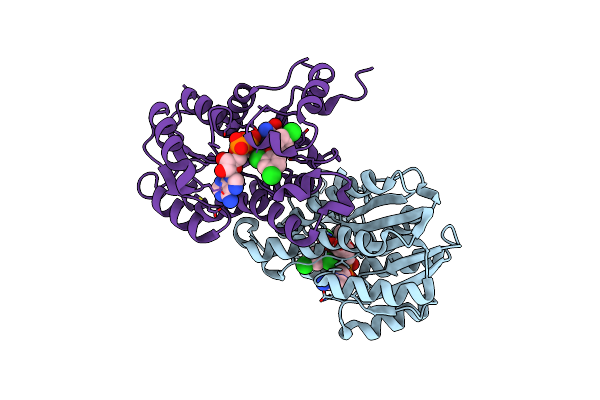 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-04-20 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, TCL |
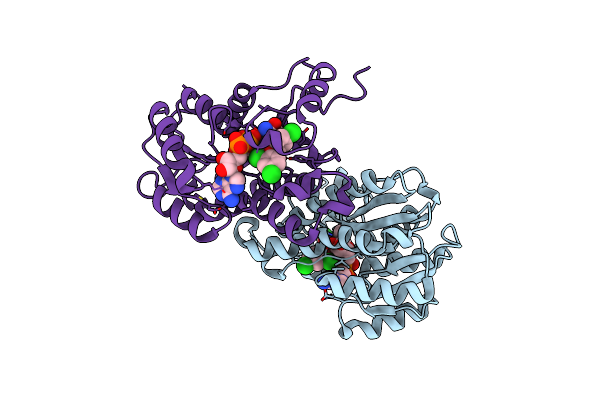 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-04-20 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, TCL |
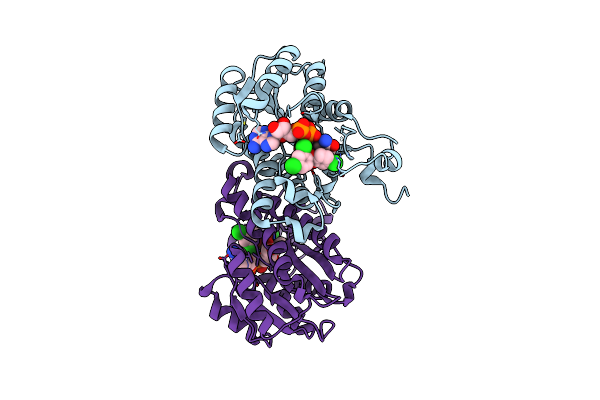 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-04-20 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, TCL |
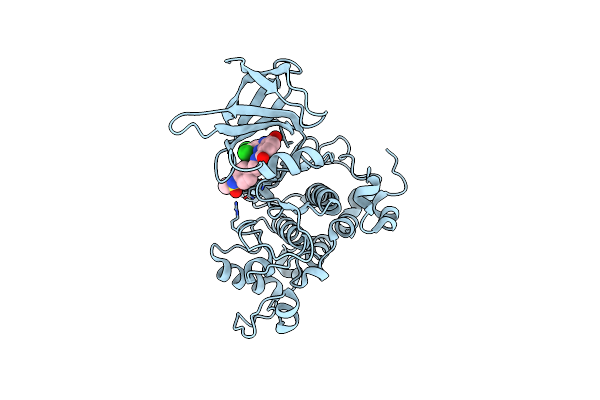 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2007-10-23 Classification: TRANSFERASE Ligands: HBM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-07-03 Classification: LIGASE |
 |
Structure Of Human Prl-3, The Phosphatase Associated With Cancer Metastasis
|
 |
Structural Basis For The Selective Inhibition Of Jnk1 By The Scaffolding Protein Jip1 And Sp600125
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2004-08-30 Classification: TRANSFERASE |
 |
Structural Basis For The Selective Inhibition Of Jnk1 By The Scaffolding Protein Jip1 And Sp600125
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-08-30 Classification: TRANSFERASE Ligands: 537 |
 |
Crystal Structure Of Human Phosphodiesterase 5 Complexed With Vardenafil(Levitra)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-07-09 Classification: HYDROLASE Ligands: ZN, MG, VDN |
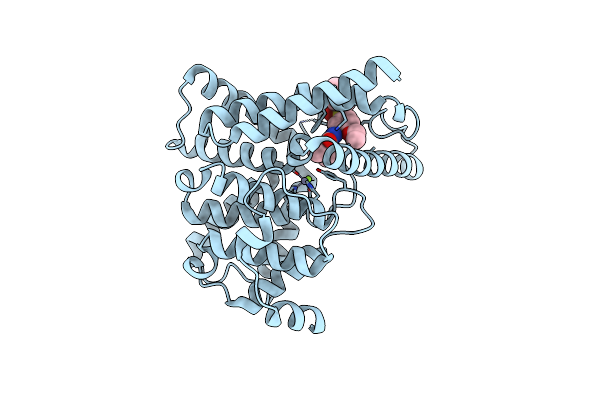 |
Crystal Structure Of Human Phosphodiesterase 5 Complexed With Sildenafil(Viagra)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-05-11 Classification: HYDROLASE Ligands: ZN, MG, VIA |
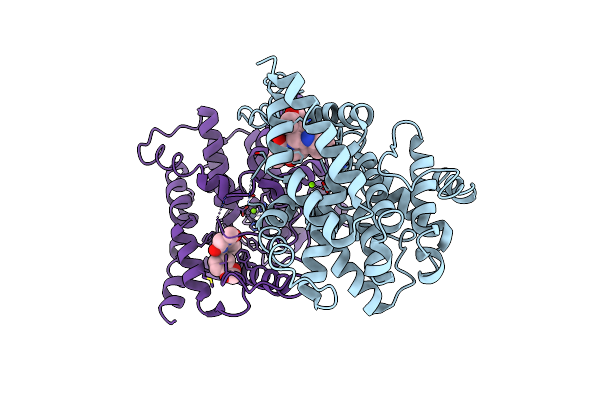 |
Crystal Structure Of Human Phosphodiesterase 5 Complexed With Tadalafil(Cialis)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2004-05-11 Classification: HYDROLASE Ligands: ZN, MG, CIA |
 |
Molecular Basis For The Local Confomational Rearrangement Of Human Phosphoserine Phosphatase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2003-04-01 Classification: HYDROLASE Ligands: APO |
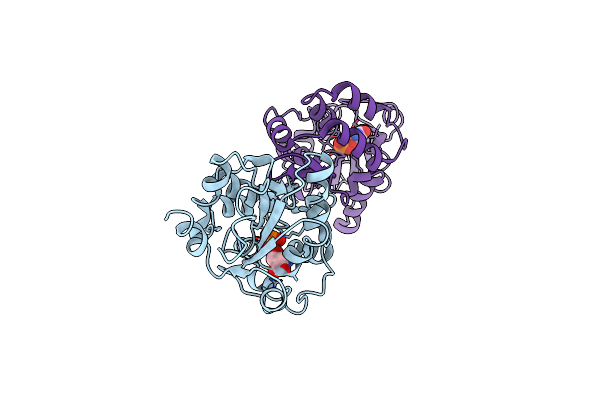 |
Molecular Basis For The Local Conformational Rearrangement Of Human Phosphoserine Phosphatase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-04-01 Classification: HYDROLASE Ligands: PO4, SER |
 |
Solution Structure And Backbone Dynamics Of A Double Mutant Single-Chain Monellin(Scm) Determined By Nuclear Magnetic Resonance Spectroscopy
Organism: Dioscoreophyllum cumminsii
Method: SOLUTION NMR Release Date: 2001-06-06 Classification: PLANT PROTEIN |
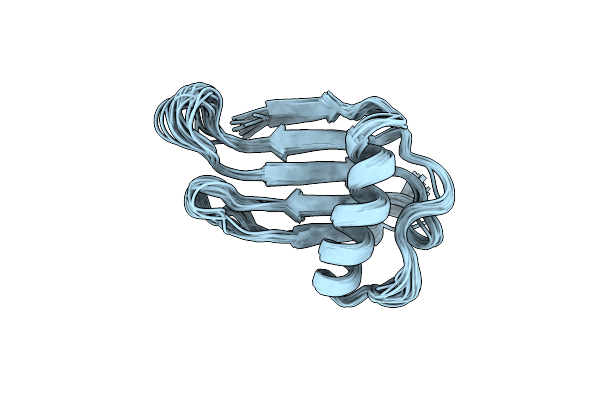 |
High-Resolution Solution Structure Of A Sweet Protein Single-Chain Monellin (Scm) Determined By Nuclear Magnetic Resonance Spectroscopy And Dynamical Simulated Annealing Calculations, 21 Structures
Organism: Dioscoreophyllum cumminsii
Method: SOLUTION NMR Release Date: 1999-06-08 Classification: SWEET PROTEIN |
 |
Two Crystal Structures Of A Potently Sweet Protein: Natural Monellin At 2.75 Angstroms Resolution And Single-Chain Monellin At 1.7 Angstroms Resolution
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1994-05-31 Classification: SWEET-TASTING PROTEIN |

