Search Count: 15
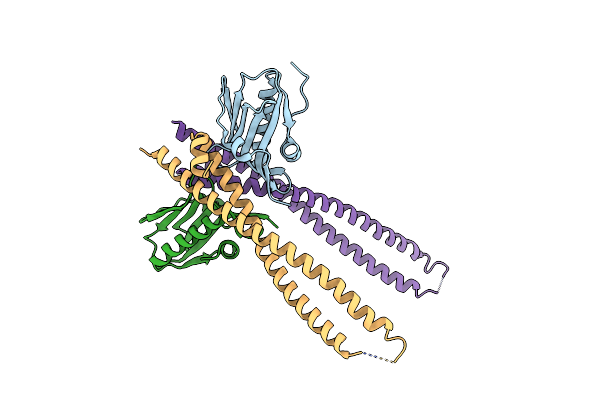 |
Crystal Structure Of Ns1-Ed Of Vietnam Influenza A Virus In Complex With The P85-Beta-Ish2 Domain Of Human Pi3K
Organism: Influenza a virus (a/viet nam/1203/2004(h5n1)), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-07-13 Classification: VIRAL PROTEIN |
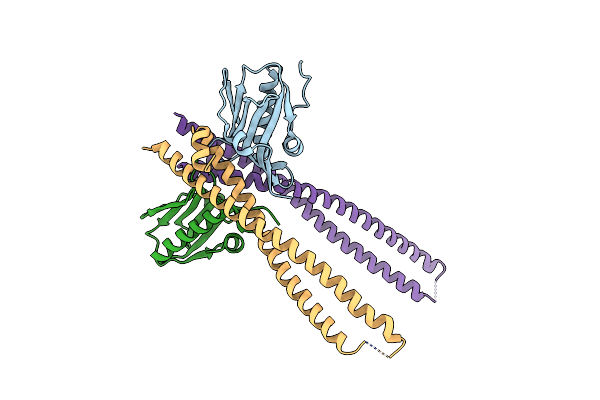 |
The Complex Of 1918 Ns1-Ed And The Ish2 Domain Of The Human P85Beta Subunit Of Pi3K
Organism: Influenza a virus (strain a/brevig mission/1/1918 h1n1), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-04-22 Classification: VIRAL PROTEIN |
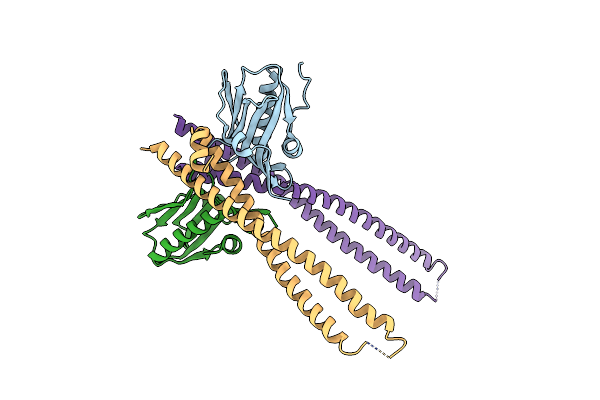 |
Crystal Structure Of 1918 Ns1-Ed W187A In Complex With The P85-Beta-Ish2 Domain Of Human Pi3K
Organism: Influenza a virus (strain a/brevig mission/1/1918 h1n1), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2020-04-22 Classification: VIRAL PROTEIN |
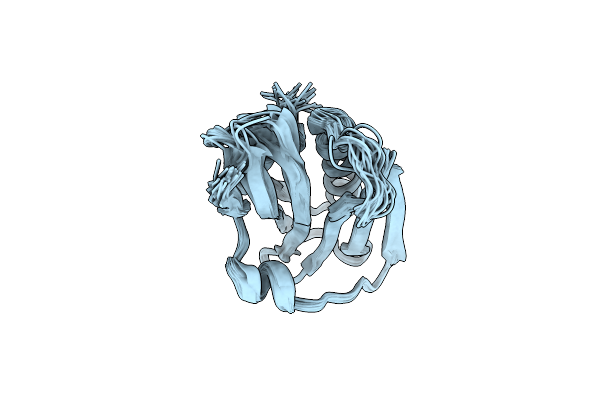 |
Organism: Influenza a virus
Method: SOLUTION NMR Release Date: 2020-01-08 Classification: VIRAL PROTEIN |
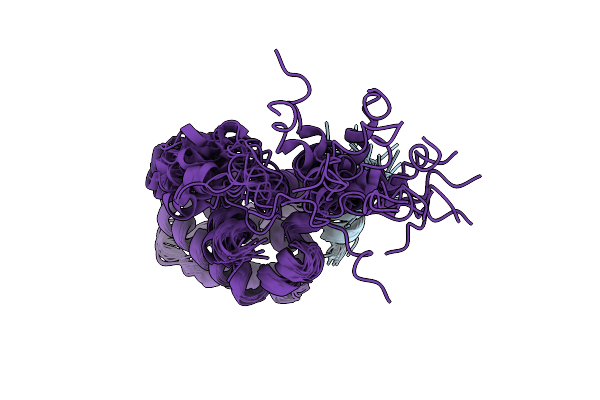 |
 |
The Molecular Mechanisms By Which Ns1 Of The 1918 Spanish Influenza A Virus Hijack Host Protein-Protein Interactions
Organism: Homo sapiens, Orthomyxoviridae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-08-08 Classification: protein binding/viral protein |
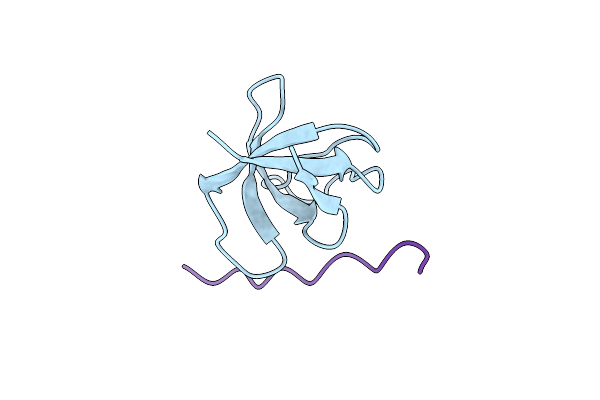 |
The Molecular Mechanisms By Which Ns1 Of The 1918 Spanish Influenza A Virus Hijack Host Protein-Protein Interactions
Organism: Homo sapiens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-08-09 Classification: VIRAL PROTEIN |
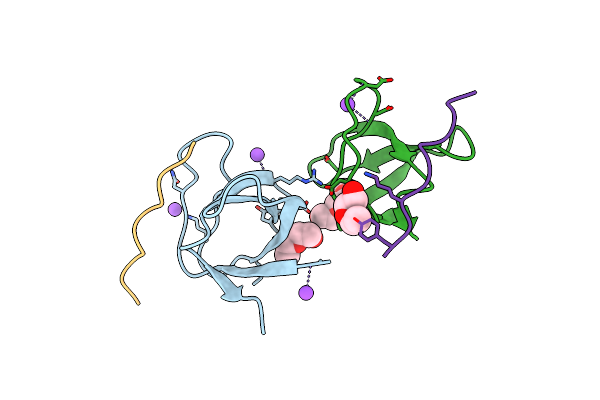 |
Structure, Thermodynamics, And The Role Of Conformational Dynamics In The Interactions Between The N-Terminal Sh3 Domain Of Crkii And Proline-Rich Motifs In Cabl
Organism: Mus musculus, Endothia gyrosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-06-29 Classification: SIGNALING PROTEIN Ligands: PEG, NA, P4G |
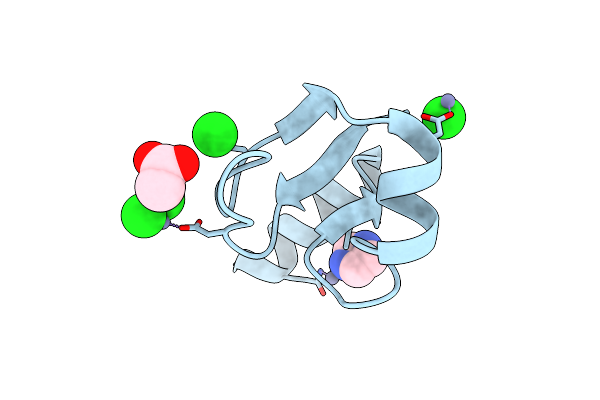 |
Crystal Structure Of N-Terminal Domain Of Ribosomal Protein L9 (Ntl9), G34Da
Method: X-RAY DIFFRACTION
Resolution:1.57 Å Release Date: 2007-06-12 Classification: STRUCTURAL PROTEIN Ligands: ZN, CL, IMD, ACY |
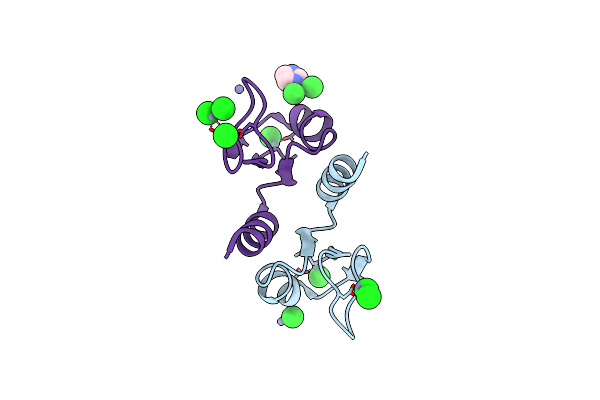 |
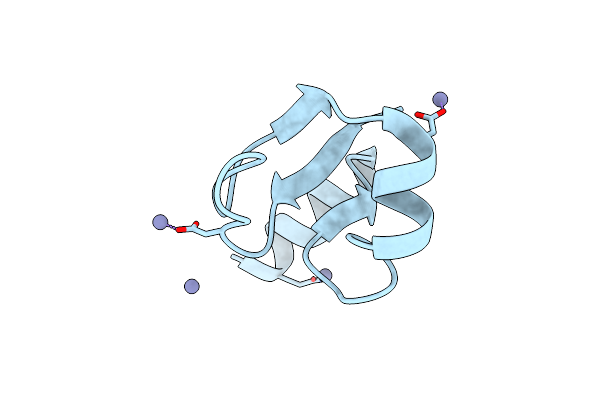
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2007-05-29 Classification: RNA BINDING PROTEIN Ligands: ZN, CL, IMD |
 |
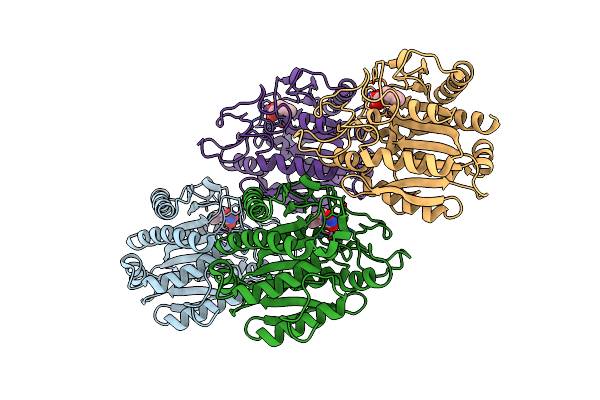
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-05-29 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
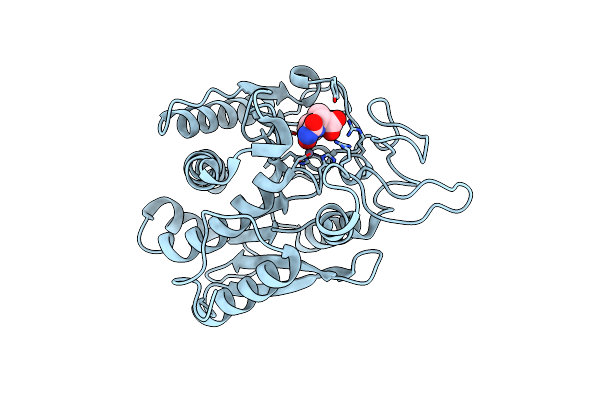
Crystal Structure Of Bovine Pancreatic Carboxypeptidase A Complexed With L-N-Hydroxyaminocarbonyl Phenylalanine At 2.3 A
Organism: Bos bovis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2001-11-23 Classification: CARBOXYPEPTIDASE Ligands: ZN, LHY |
 |
Crystal Structure Of Bovine Pancreatic Carboxypeptidase A Complexed With D-N-Hydroxyaminocarbonyl Phenylalanine At 2.3 A
Organism: Bos bovis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2001-11-15 Classification: CARBOXYPEPTIDASE Ligands: INF, ZN |
 |
Crystal Structure Of Bovine Pancreatic Carboxypeptidase A Complexed With Aminocarbonylphenylalanine At 1.75 A
Organism: Bos bovis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2001-11-15 Classification: CARBOXYPEPTIDASE Ligands: ZN, ING |
 |

