Search Count: 302
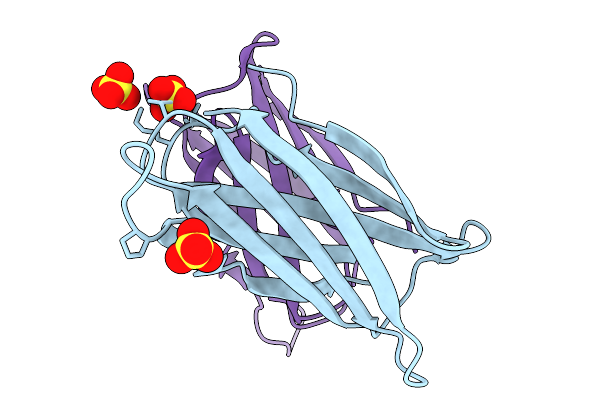 |
Crystal Structure Of The Helicobacter Pylori Copper Resistance Determinant Crda In The Apo Form
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: SO4 |
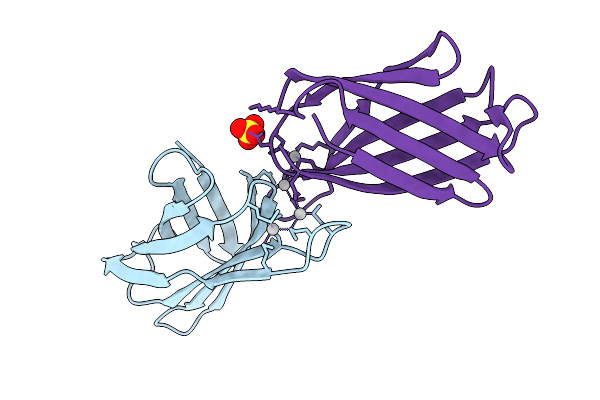 |
Crystal Structure Of The Helicobacter Pylori Copper Resistance Determinant Crda In Complex With Silver Ions In Space Group P21
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: AG, SO4 |
 |
Crystal Structure Of The Helicobacter Pylori Copper Resistance Determinant Crda In Complex With Silver Ions In Space Group P1
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: AG, GOL |
 |
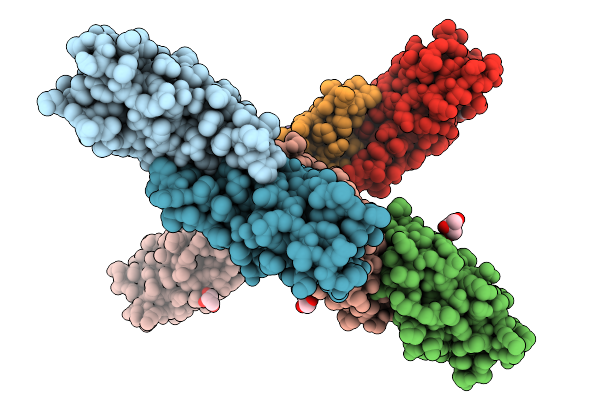
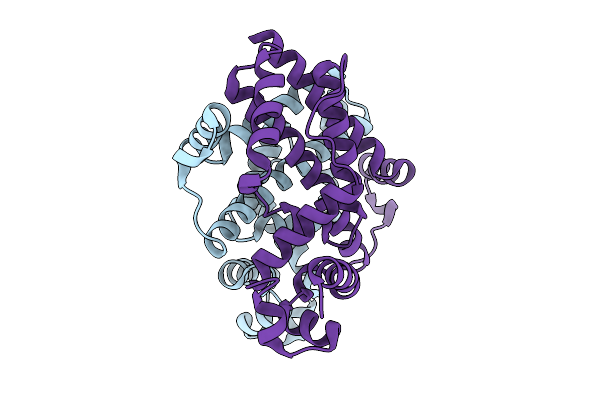
Crystal Structure Of The Engineered Sulfonylurea Repressor Esr (L7-D1), Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN |
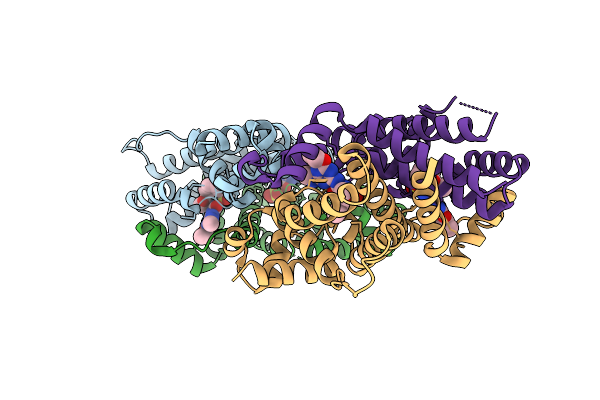 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Esr (L11-C6), Bound To Ethametsulfuron-Methyl
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: RXF |
 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Csr (L4.2-20), Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN |
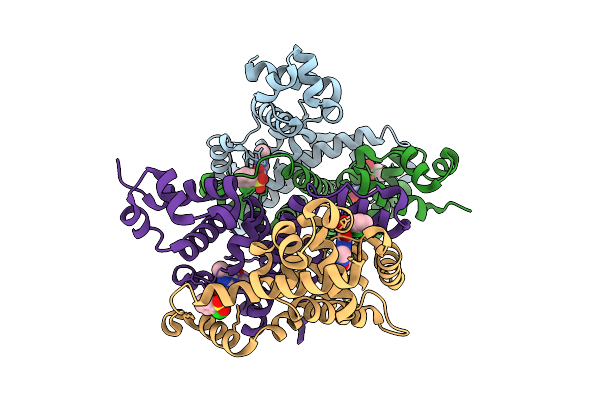 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Csr (L4.2-20), Bound To Chlorsulfuron
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: 1CS |
 |
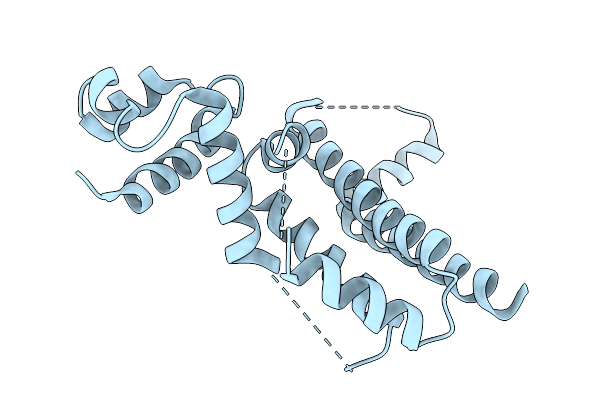
Crystal Structure Of The Moae-Like Domain Within Rv3323C From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE Ligands: MES, GOL |
 |
Cryo-Em Structure Of Outward State Anhydromuropeptide Permease (Ampg) Complex With Glcnac-1,6-Anhmurnac
Organism: Homo sapiens, Synthetic construct, Yokenella regensburgei, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: 2YP |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: ADP, ATP |
 |
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSFERASE |
 |
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: ACO, PO4 |
 |
Organism: Yokenella regensburgei
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Inward-Facing Anhydromuropeptide Permease (Ampg) In Complex With Glcnac-1,6-Anhmurnac
Organism: Yokenella regensburgei
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: 2YP |
 |
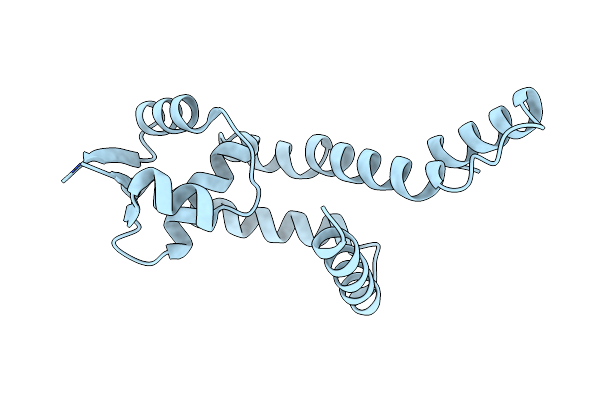
Cryo-Em Structure Of Outward State Anhydromuropeptide Permease (Ampg) G50W/L269W
Organism: Yokenella regensburgei, Escherichia coli, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: PEPTIDE BINDING PROTEIN |
 |
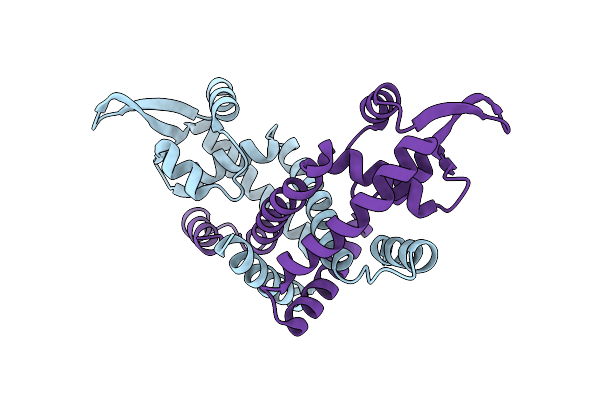
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-11-13 Classification: TRANSCRIPTION |
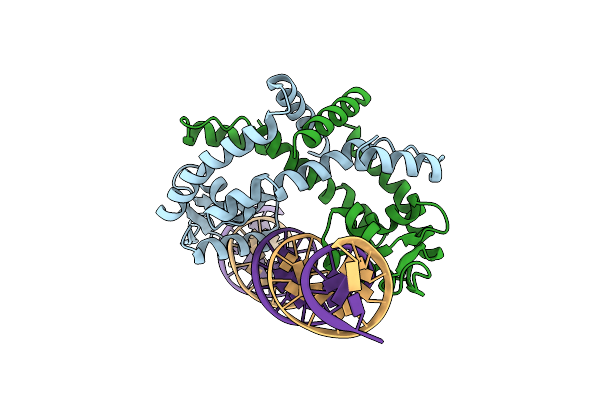 |
Crystal Structure Of Burkholderia Thailandensis Mftr In Complex With Operator Dna
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-13 Classification: TRANSCRIPTION |
 |
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-11-13 Classification: TRANSCRIPTION Ligands: CL |
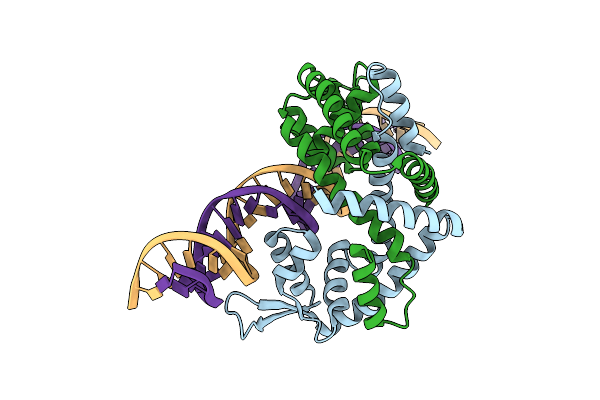 |
Crystal Structure Of Pectobacterium Atrosepticum Pecs In Complex With Operator Dna
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-11-13 Classification: TRANSCRIPTION |
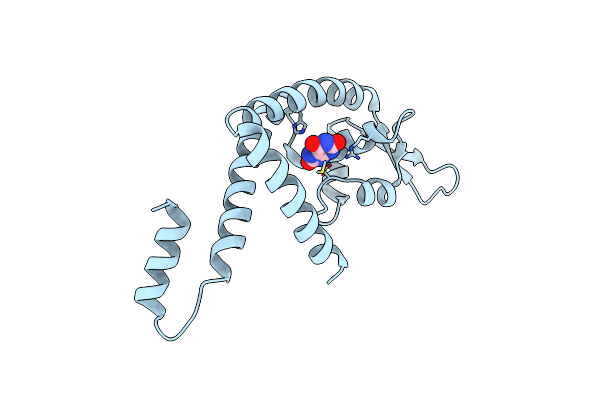 |
Crystal Structure Of Pectobacterium Atrosepticum Pecs In Complex With Uric Acid
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-11-13 Classification: TRANSCRIPTION Ligands: URC |

