Search Count: 265
 |
Structure Of A Bacterial Serpin Choloropin Derived From Cholorobium Limicola
Organism: Chlorobium limicola (strain dsm 245 / nbrc 103803 / 6330)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-27 Classification: DNA BINDING PROTEIN Ligands: ZN, GOL |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: MG, CL, EDO, CA, AVJ, ZN |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: AVJ, MG, CL |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: AVJ, MG, CL, PEG |
 |
The Structure Of Eanb/Y353F-Cys412-Persulfide In Tetrahedral Intermediate State With Ergothioneine
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: CL, MG, BR, PGE, LW8 |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: AVJ, MG, CL, GOL, EDO, PEG, PG4, PDO, IMD, NI |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, AVJ, CL, PGE, GOL, IMD, NA, EDO |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, CL, PEG, MPO, EDO, PGE |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, AVJ, PEG, EDO, CL, BR |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, LW8, CL, EDO |
 |
The Structure Of Eanb/Y353A Complex With Ergothioneine Covalent Linked With Persulfide Cys412
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: DV6, MG, CL, GOL, IMD, ZN, BR, PEG, EDO |
 |
Structural Determination Of The Carboxy-Terminal Portion Of Atp-Citrate Lyase
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-12-18 Classification: LYASE Ligands: ACO, PO4, GOL, NA |
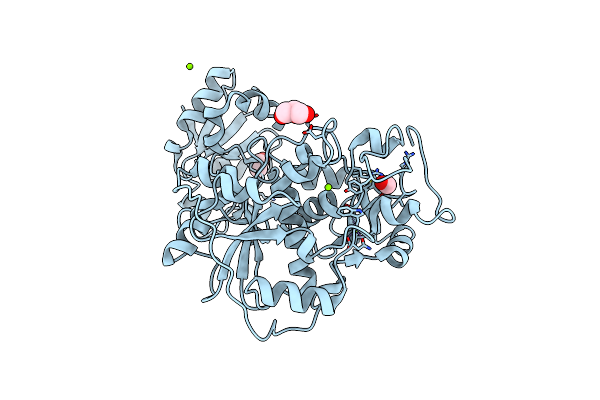 |
Native Crystal Structure Of Anaerobic Ergothioneine Biosynthesis Enzyme From Chlorobium Limicola.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: MG, CL, FMT, EOH, GOL |
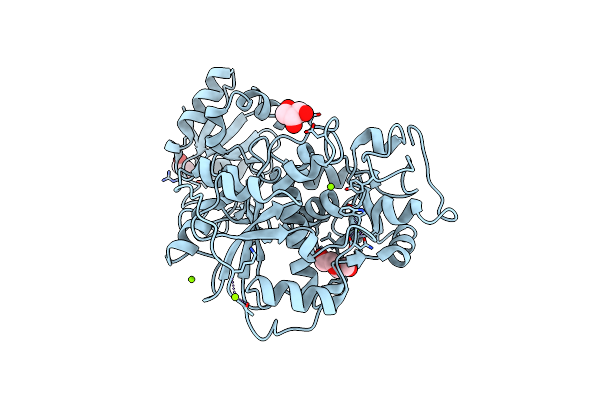 |
Crystal Structure Of Anaerobic Ergothioneine Biosynthesis Enzyme From Chlorobium Limicola In Persulfide Form.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: MG, CL, EDO, PEG, GOL |
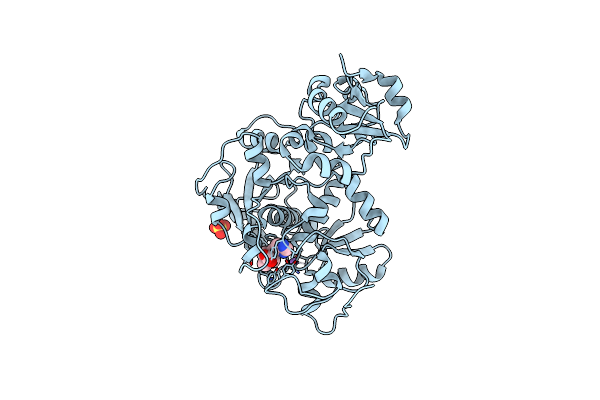 |
Crystal Structure Of Anaerobic Ergothioneine Biosynthesis Enzyme From Chlorobium Limicola In Complex With Natural Substrate Trimethyl Histidine.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: AVJ, NA, CL, SO4 |
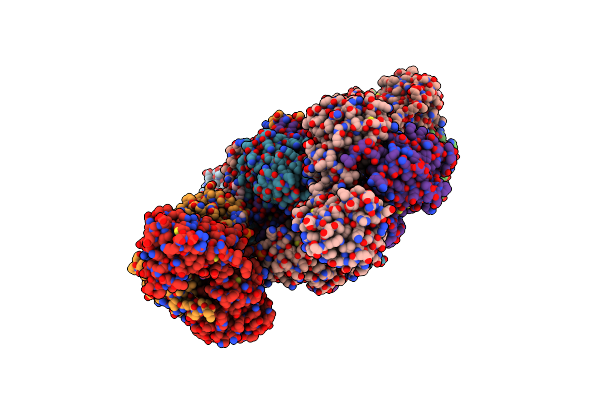 |
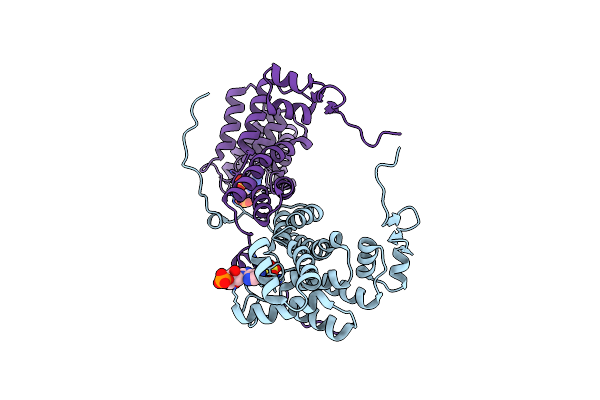
Structure Of Atp Citrate Lyase From Chlorobium Limicola In Complex With Citrate And Coenzyme A.
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2019-04-10 Classification: LYASE Ligands: FLC, COA, PGE, TRS, PG4 |
 |
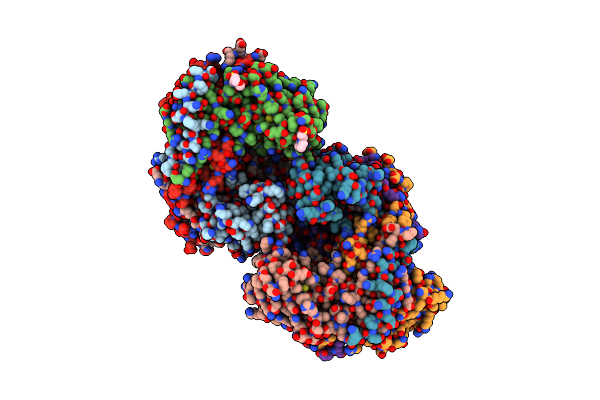
Structure Of The Citryl-Coa Lyase Core Module Of Chlorobium Limicola Atp Citrate Lyase (Space Group P3121)
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-04-10 Classification: LYASE Ligands: COA, SO4 |
 |
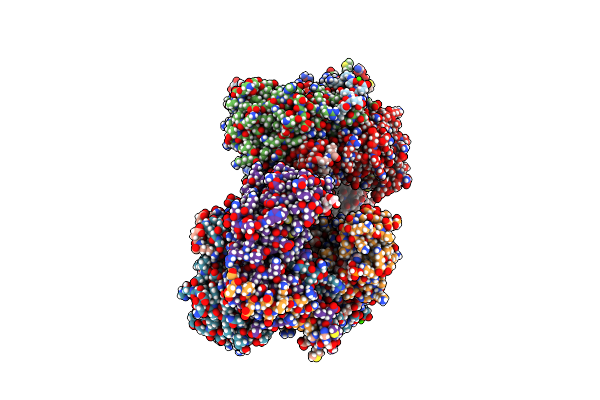
Structure Of The Citryl-Coa Lyase Core Module Of Chlorobium Limicola Atp Citrate Lyase (Space Group P21)
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-04-10 Classification: LYASE Ligands: FLC, MES |
 |
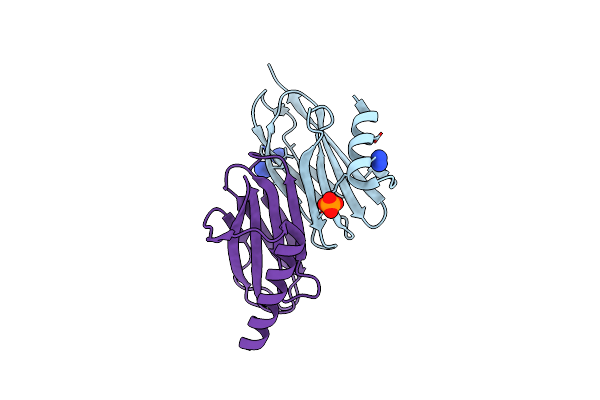
Citryl-Coa Lyase Core Module Of Chlorobium Limicola Atp Citrate Lyase In Complex With Acetyl-Coa And L-Malate
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-04-10 Classification: LYASE Ligands: CA, LMR, ACO, PG4, PGE |
 |
Structure Of The Sulfur Carrier Protein Soxy From Chlorobium Limicola F Thiosulfatophilum
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2007-03-13 Classification: LIGAND BINDING PROTEIN Ligands: CL, PO4, HDZ |

