Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: DE NOVO PROTEIN Ligands: RET, SO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
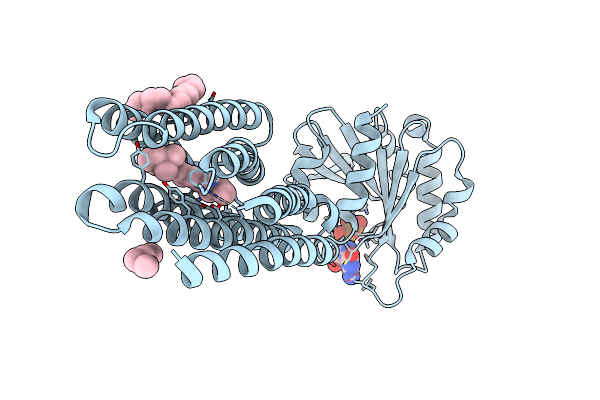
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
 |
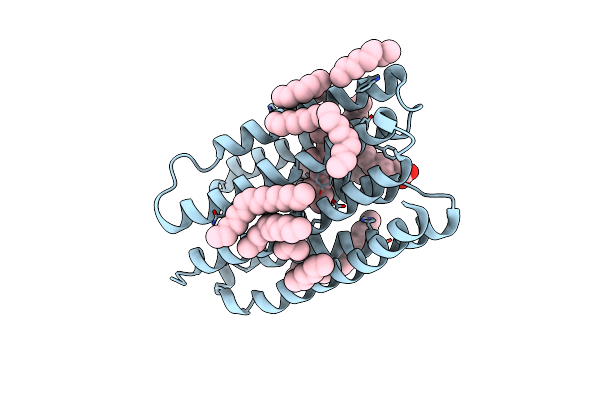
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
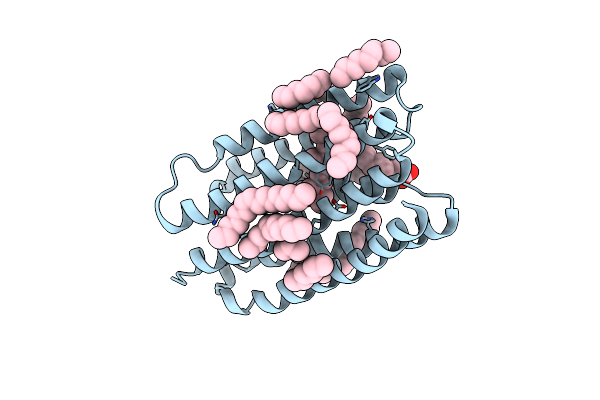
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
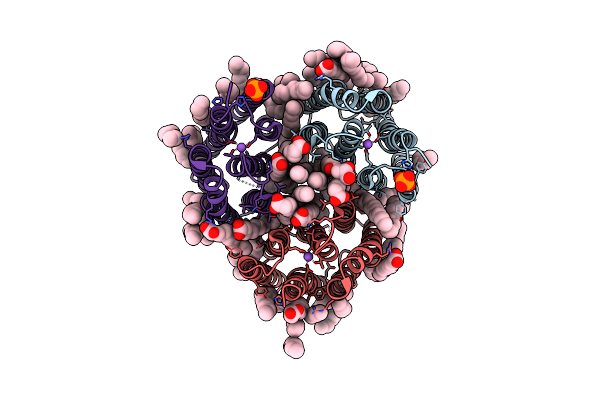
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
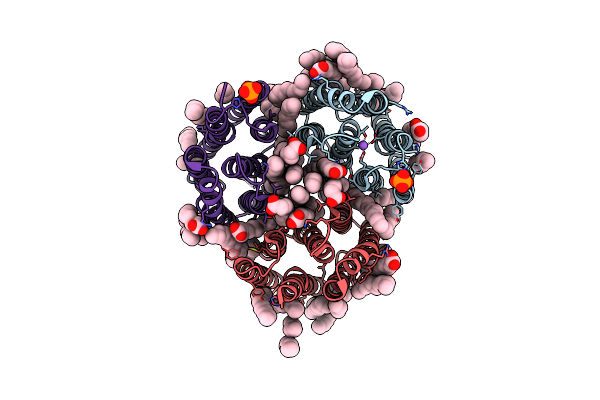
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 8.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, NA, PO4 |
 |
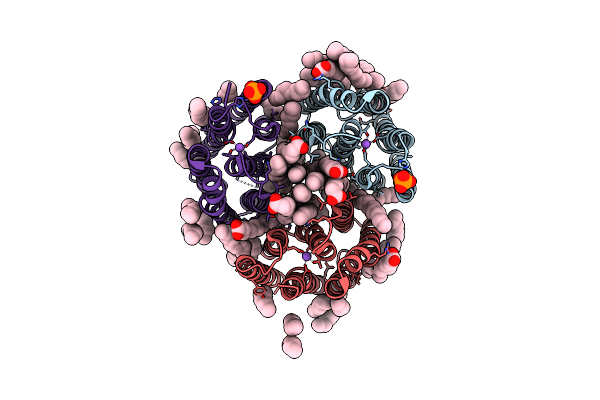
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 8.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4, NA |
 |
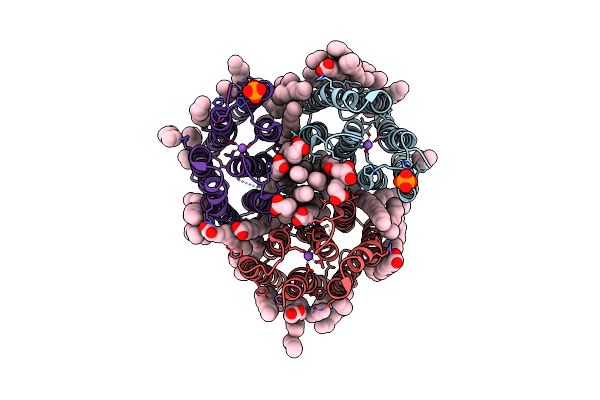
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The L State At Ph 8.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, NA, PO4 |
 |
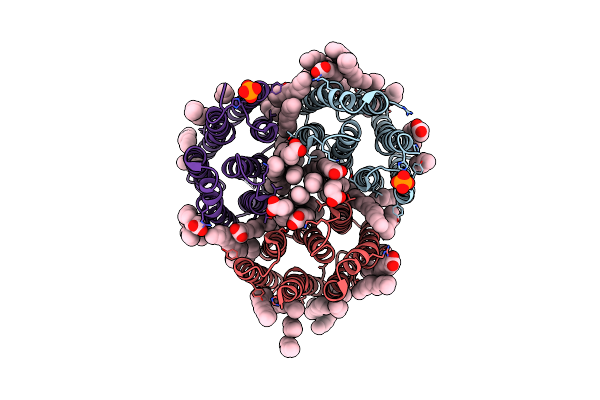
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 7.0 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, NA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 7.0 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 5.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: OLA, OLC, LFA, PO4, NA |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 5.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 7.6 In The Absence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 7.6 In The Absence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 8.2 At Room Temperature, 7.5-Ms-Long Snapshots
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 8.2 At Room Temperature, 500-Mks-Long Snapshots
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Activated State At Ph 8.2 At Room Temperature, 250-750-Mks-Snapshot
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |