Search Count: 41
 |
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: VIRAL PROTEIN Ligands: ZN, SAH, A1JDF, CL, EDO, IMD, IPA, SO4 |
 |
Crystal Structure Of The Artificial Protein Metp In Complex With Cadmium Ion At Different Temperature (Data Set At 100 K)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2024-12-04 Classification: DE NOVO PROTEIN Ligands: CD |
 |
Crystal Structure Of The Artificial Protein Metp In Complex With Cadmium Ion At Different Temperature, 160 K
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-04 Classification: DE NOVO PROTEIN Ligands: CD |
 |
Crystal Structure Of The Artificial Protein Metp In Complex With Cadmium Ion At Different Temperature, 130 K
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-04 Classification: DE NOVO PROTEIN Ligands: CD |
 |
Crystal Structure Of The Artificial Protein Metp In Complex With Cadmium Ion At Different Temperatures. 200 K Data Collection
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-04 Classification: DE NOVO PROTEIN Ligands: CD |
 |
Crystal Structure Of The Artificial Protein Metp In Complex With Cadmium Ion At Different Temperatures. Room Temperature Data Collection
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-04 Classification: DE NOVO PROTEIN Ligands: CD |
 |
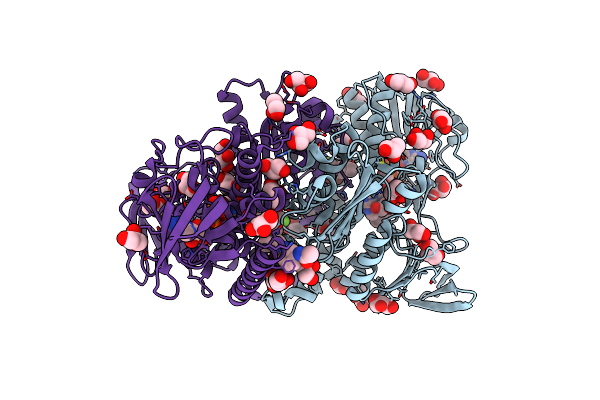
Cryo-Em Structure Of Cell-Free Synthesized Human Histamine H2 Receptor Coupled To Heterotrimeric Gs Protein In Lipid Environment
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: MEMBRANE PROTEIN Ligands: HSM |
 |
Co-Crystal Structure Of Optimized Analog Tdi-13537 Provided New Insights Into The Potency Determinants Of The Sulfonamide Inhibitor Series
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-01-03 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, U4I, GOL |
 |
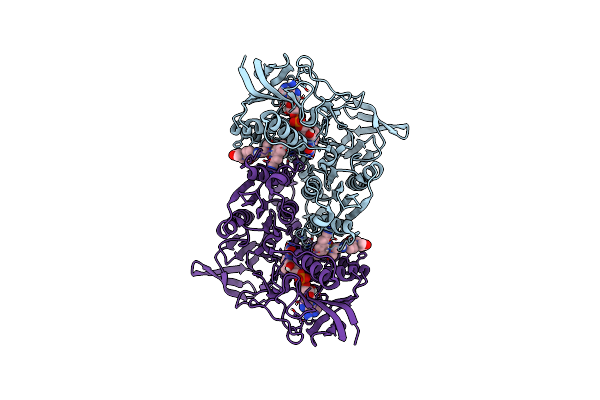
The Crystal Structure Of Metp In Complex With Zn At A Resolution Of 1.34 A.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2023-02-15 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-02-15 Classification: DE NOVO PROTEIN Ligands: CO |
 |
The Crystal Structure Of Metp In Complex With Cd At A Resolution Of 1.29 A.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2023-02-15 Classification: DE NOVO PROTEIN Ligands: CD |
 |
Cryo-Em Structure Of Mtb Lpd Bound To Inhibitor Complex With 2-((2-Cyano-N,5-Dimethyl-1H-Indole)-7-Sulfonamido)-N-(4-(Oxetan-3-Yl)-3,4-Dihydro-2H-Benzo[B] [1,4]Oxazin-7-Yl)Acetamide
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: FAD, OU6 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-10-06 Classification: SIGNALING PROTEIN Ligands: 1S2, ACT, DMS, EDO |
 |
Macrocyclic Peptides Tdi5575 That Selectively Inhibit The Mycobacterium Tuberculosis Proteasome
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-04-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: U5Y, CIT, DMF |
 |
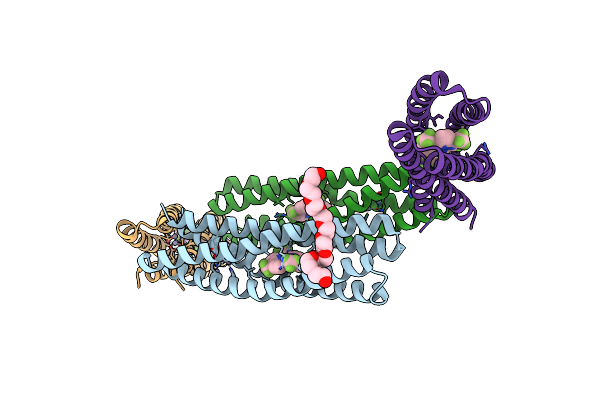
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2021-01-27 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FAD, WPM, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-12-09 Classification: DE NOVO PROTEIN Ligands: ZN, 7BU, 2PE |
 |
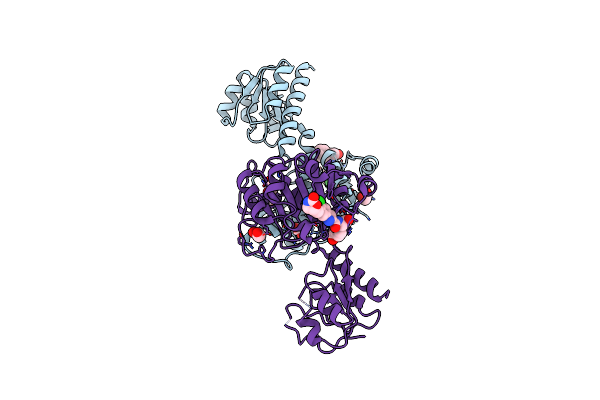
Crystal Structure Of Mycobacterium Tuberculosis Proteasome In Complex With Phenylimidazole-Based Inhibitor A85
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-10-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DMF, M6M, CIT |
 |
Crystal Structure Of Mycobacterium Tuberculosis Proteasome In Complex With Phenylimidazole-Based Inhibitor A86
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-10-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DMF, M6Y, CIT |
 |
Crystal Structure Of Mycobacterium Tuberculosis Proteasome In Complex With Phenylimidazole-Based Inhibitor B6
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-10-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: M9G |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-07-24 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: ONV, EDO |

