Search Count: 52
 |
Organism: Aequorea victoria, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: SIGNALING PROTEIN |
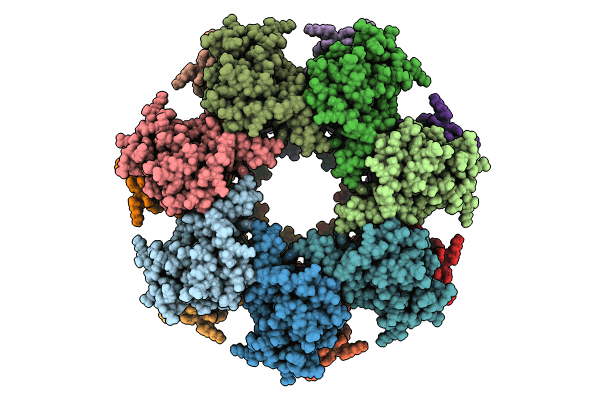 |
Cryo-Em Structure Of Tetradecameric Camkii Beta Holoenzyme T287A T306A T307A
Organism: Aequorea victoria, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: SIGNALING PROTEIN |
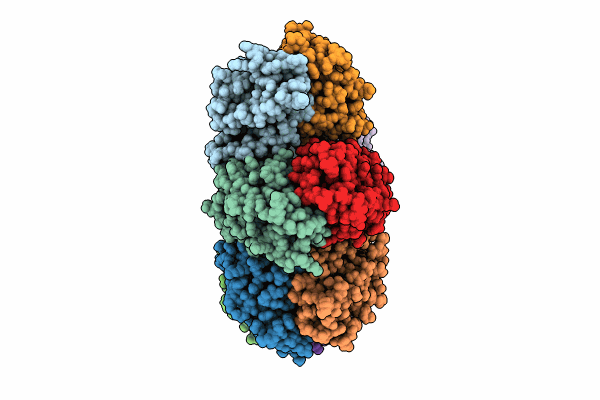 |
Organism: Aequorea victoria, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: SIGNALING PROTEIN |
 |
Organism: Aequorea victoria, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: SIGNALING PROTEIN |
 |
Organism: Aequorea victoria, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: SIGNALING PROTEIN |
 |
Organism: Aequorea victoria, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: SIGNALING PROTEIN |
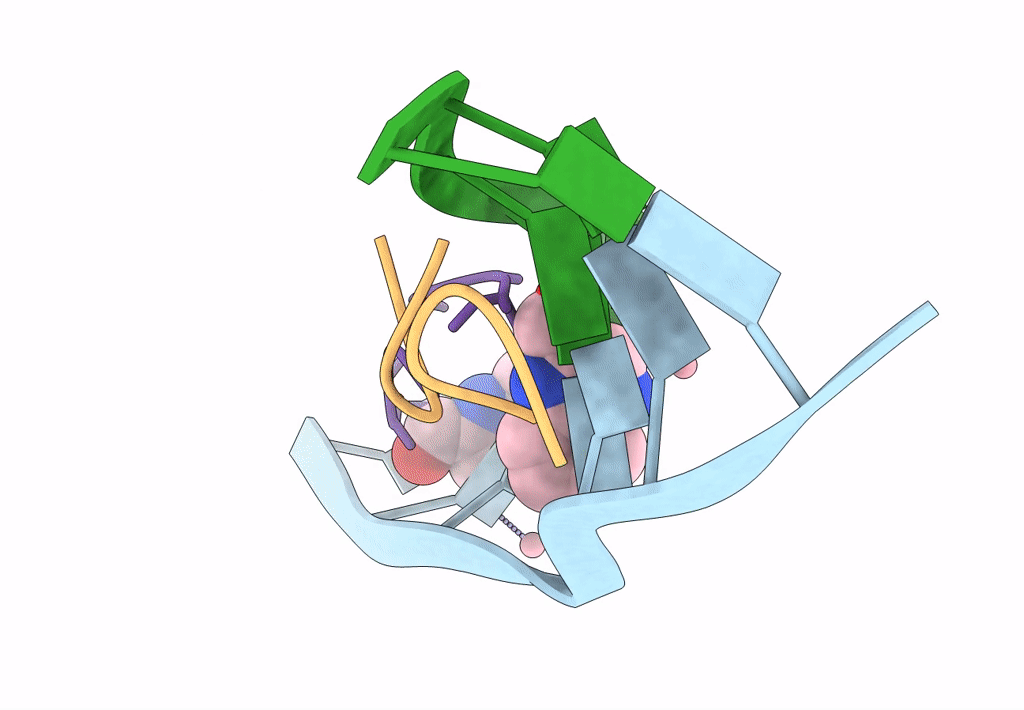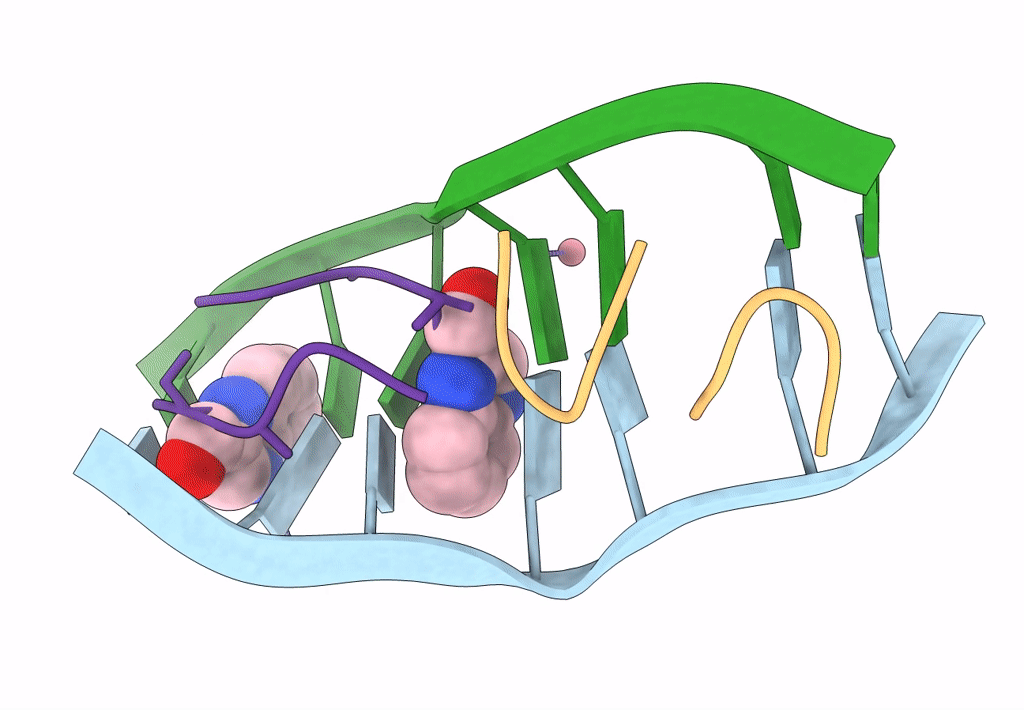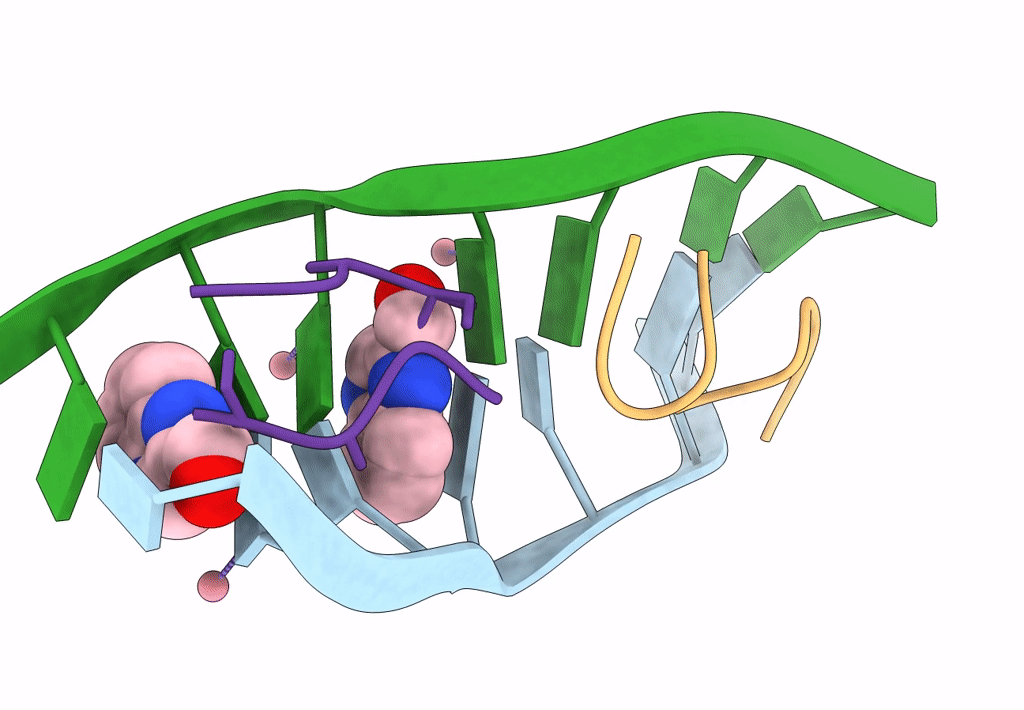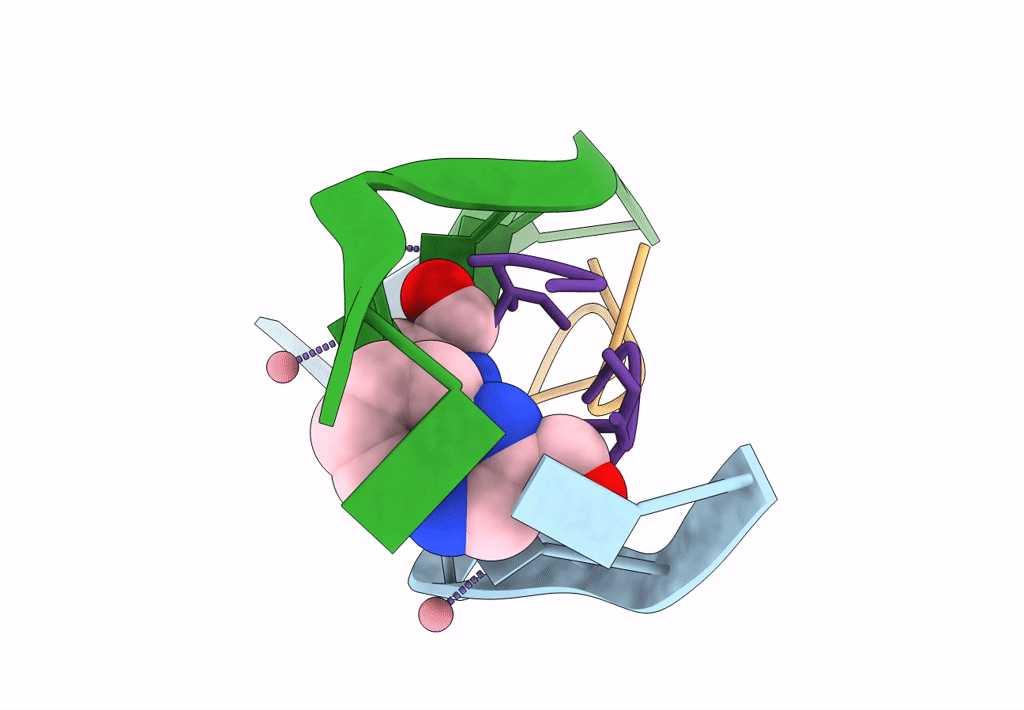 |
Organism: Synthetic construct, Streptomyces
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-12-14 Classification: DNA/ANTIBIOTIC Ligands: CO, QUI |
 |
Organism: Synthetic construct, Streptomyces
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-12-14 Classification: DNA/ANTIBIOTIC Ligands: ZN, K, CL, QUI |
 |
Organism: Synthetic construct, Streptomyces
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2022-12-14 Classification: DNA/ANTIBIOTIC Ligands: QUI, MN |
 |
Organism: Synthetic construct, Streptomyces
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2022-12-14 Classification: DNA/ANTIBIOTIC Ligands: CL, MG, QUI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: HMT |
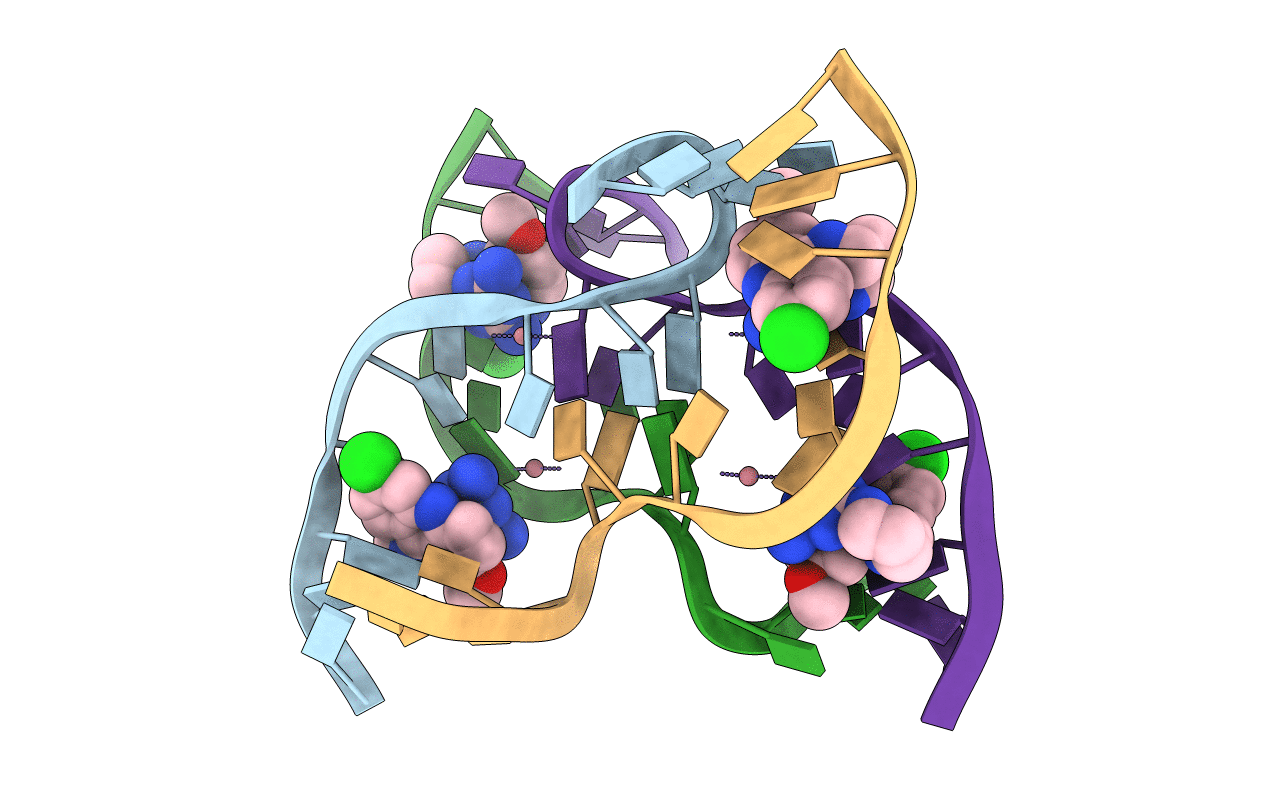 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-07-15 Classification: DNA Ligands: CO, F1R |
 |
U Shaped Head To Head Four-Way Junction In D(Ttctgctgctgaa/Ttctgcagctgaa) Sequence
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-07-15 Classification: DNA Ligands: CO, F1R |
 |
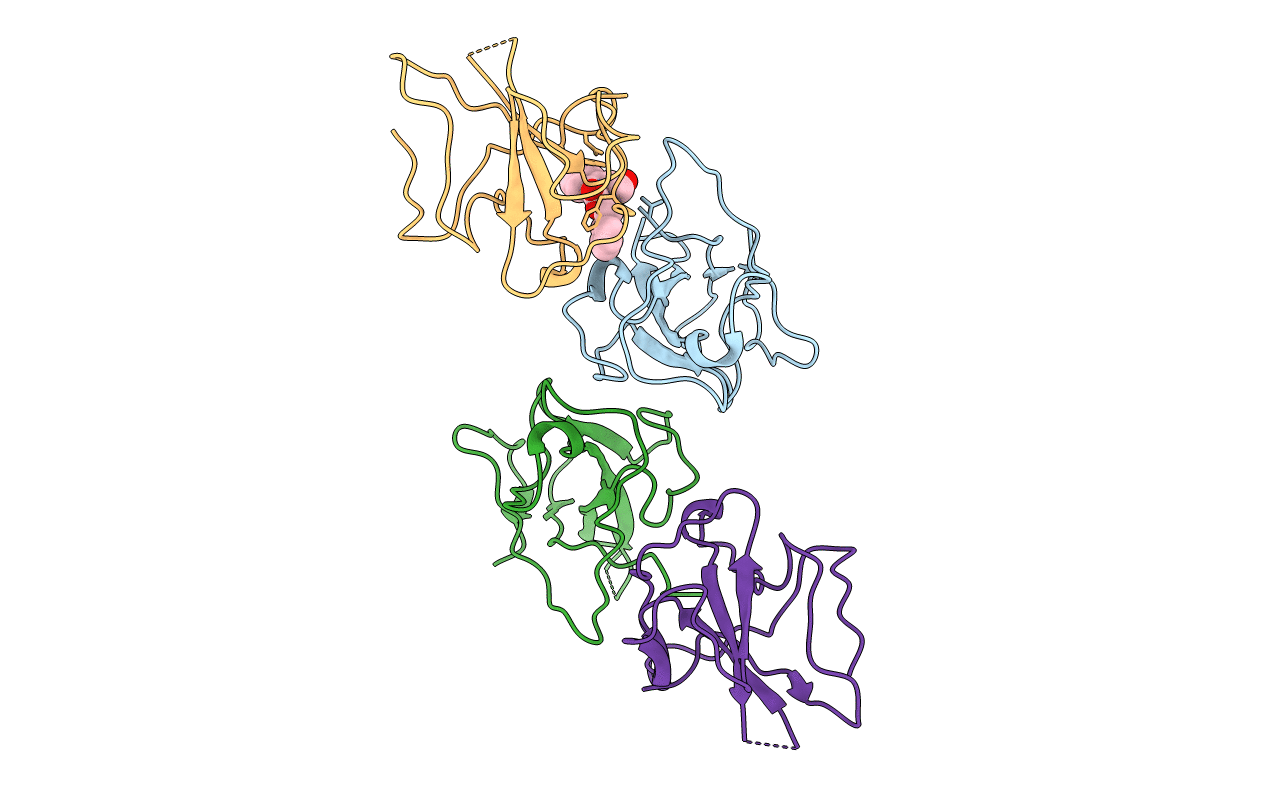
Structure Of The N-Terminal Domain Of Middle East Respiratory Syndrome Coronavirus Nucleocapsid Protein
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2020-03-25 Classification: VIRAL PROTEIN |
 |
Structure Of The N-Terminal Domain Of Middle East Respiratory Syndrome Coronavirus Nucleocapsid Protein Complexed With Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2020-03-25 Classification: VIRAL PROTEIN Ligands: DJO |
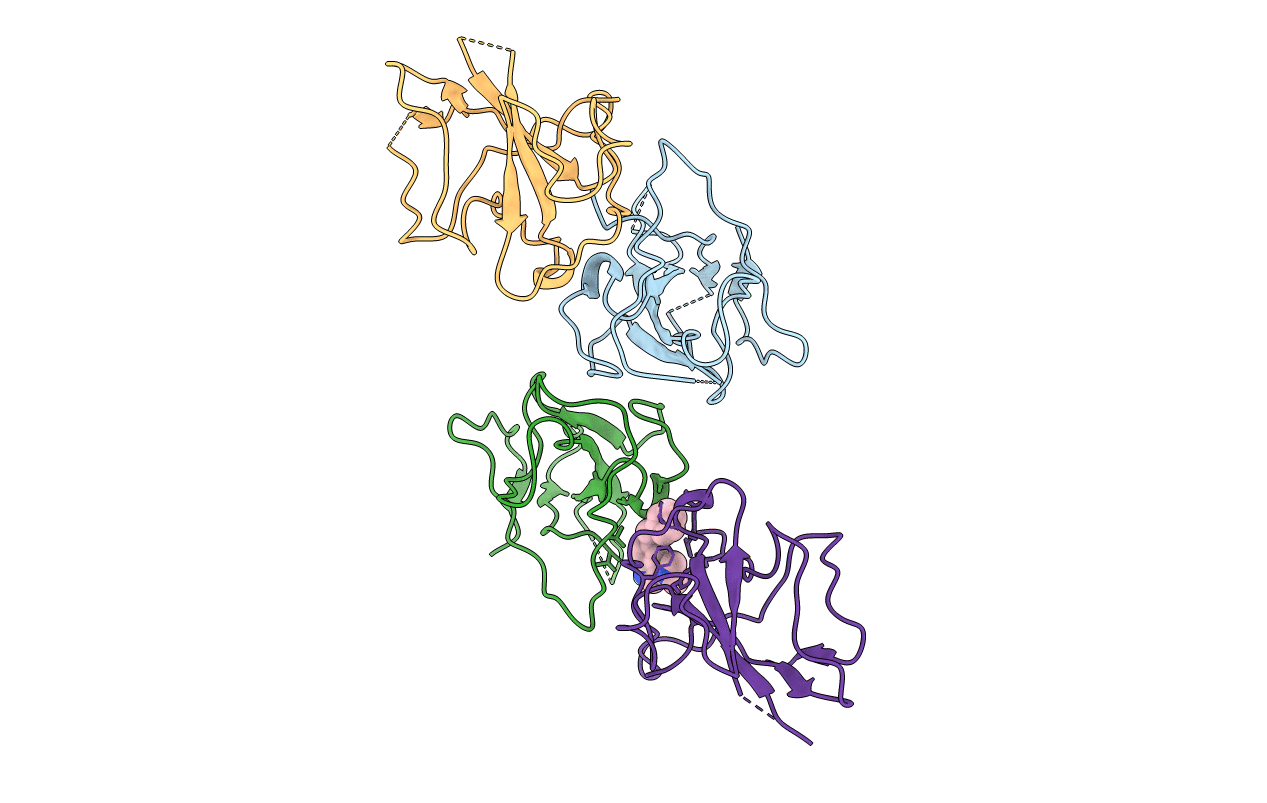 |
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2020-03-25 Classification: VIRAL PROTEIN Ligands: DJU |
 |
Organism: Pholiota squarrosa
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2020-03-04 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of A Mini Fungal Lectin, Phosl In Complex With Core-Fucosylated Chitobiose
Organism: Pholiota squarrosa
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2020-03-04 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
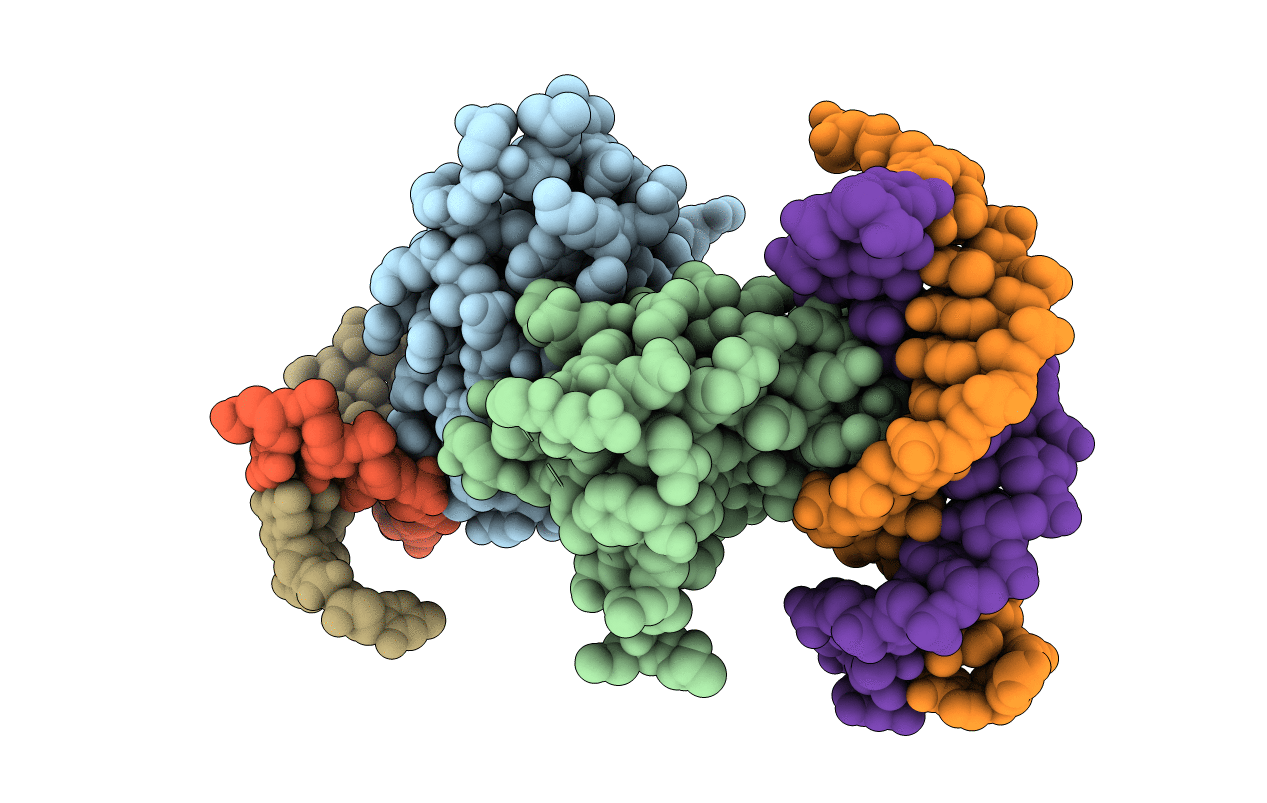 |
Crystal Structure Of Klebsiella Pneumoniae Sigma4 Of Sigmas Fusing With The Rna Polymerase Beta-Flap-Tip-Helix In Complex With -35 Element Dna
Organism: Escherichia coli, Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:3.75 Å Release Date: 2019-09-11 Classification: TRANSCRIPTION |
 |
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2019-06-05 Classification: VIRAL PROTEIN Ligands: AMP |

