Search Count: 32
 |
Organism: Stenotrophomonas sp. kctc 12332
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2018-09-05 Classification: HYDROLASE |
 |
Structural Characterization Of The Full-Length Response Regulator Spr1814 In Complex With A Phosphate Analogue Reveals A Novel Conformational Plasticity Of The Linker Region
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-04-27 Classification: DNA BINDING PROTEIN Ligands: MG, BEF |
 |
Structure Of The Full-Length Response Regulator Spr1814 In Complex With A Phosphate Analogue And B3C
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-04-27 Classification: DNA BINDING PROTEIN Ligands: MG, BEF, 4QT |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2015-07-08 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Crystal Structure Of Nitroalkane Oxidase From Pseudomonas Aeruginosa In Mutant Complex Form
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-07-08 Classification: OXIDOREDUCTASE Ligands: FMN, NIE |
 |
Crystal Structure Of Nitroalkane Oxidase From Pseudomonas Aeruginosa In Mutant Complex Form
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-07-08 Classification: OXIDOREDUCTASE Ligands: FMN, N1P |
 |
Structural Insight Into The Substrate Inhibition Mechanism Of Nadp+-Dependent Succinic Semialdehyde Dehydrogenase From Streptococcus Pyogenes
Organism: Streptococcus pyogenes mgas1882
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: SSN, SO4 |
 |
Structural Insight Into The Substrate Inhibition Mechanism Of Nadp+-Dependent Succinic Semialdehyde Dehydrogenase From Streptococcus Pyogenes
Organism: Streptococcus pyogenes mgas1882
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: SSN, SO4 |
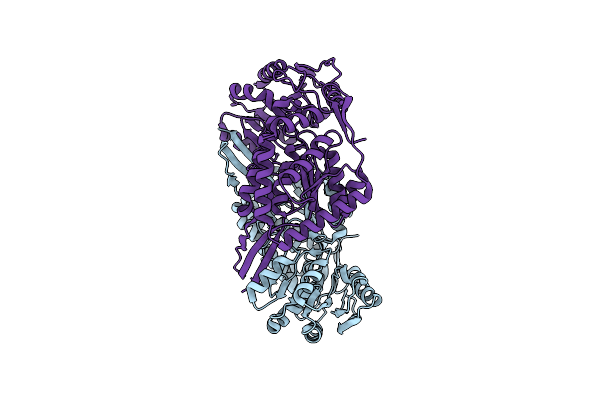 |
Crystal Structure Of Succinic Semialdehyde Dehydrogenase From Streptococcus Pyogenes In Complex With Nadp+ As The Cofactor
Organism: Streptococcus pyogenes mgas1882
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-12-10 Classification: OXIDOREDUCTASE |
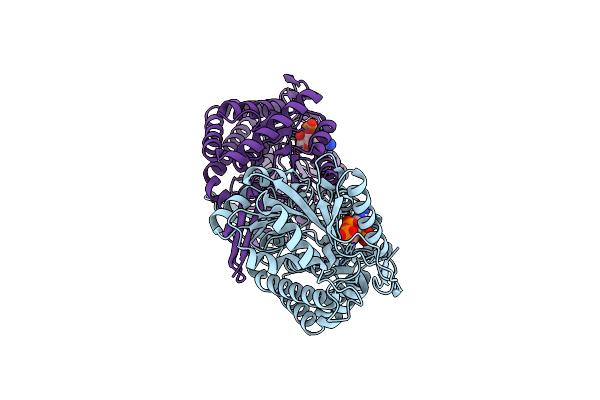 |
Crystal Structure Of Succinic Semialdehyde Dehydrogenase From Streptococcus Pyogenes In Complex With Nadp+ As The Cofactor
Organism: Streptococcus pyogenes mgas1882
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-12-10 Classification: OXIDOREDUCTASE Ligands: NAP |
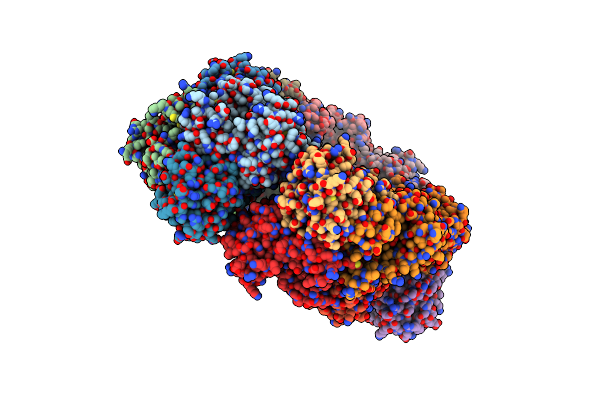 |
Crystal Structure Of The Pseudomonas Aeruginosa Enoyl-Acyl Carrier Protein Reductase (Fabi) In Apo Form
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-11-26 Classification: OXIDOREDUCTASE |
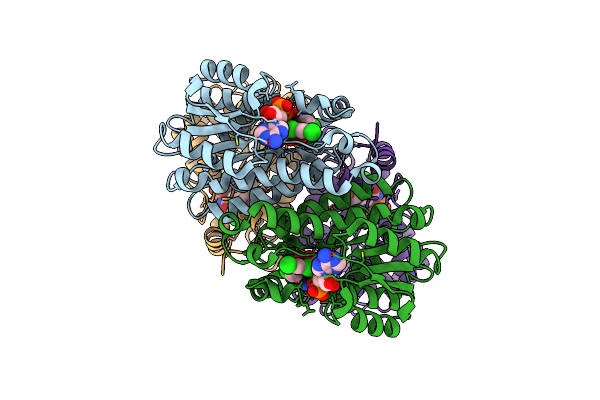 |
Crystal Structure Of The Pseudomonas Aeruginosa Enoyl-Acyl Carrier Protein Reductase (Fabi) In Complex With Nad+ And Triclosan
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-11-26 Classification: OXIDOREDUCTASE Ligands: NAD, TCL |
 |
Crystal Structure Of A Response Regulator Spr1814 From Streptococcus Pneumoniae Reveals Unique Interdomain Contacts Among Narl Family Proteins
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-06-05 Classification: TRANSCRIPTION ACTIVATOR |
 |
Crystal Structure Of Receiver Domain Of Putative Narl Family Response Regulator Spr1814 From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-05-23 Classification: TRANSCRIPTION REGULATOR Ligands: MG |
 |
Crystal Structure Of Receiver Domain Of Putative Narl Family Response Regulator Spr1814 From Streptococcus Pneumoniae In The Presence Of The Phosphoryl Analog Beryllofluoride
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2012-05-23 Classification: TRANSCRIPTION REGULATOR Ligands: MG, BEF |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-04-11 Classification: OXIDOREDUCTASE Ligands: SME |
 |
Crystal Structure Of A Glycosylated Ice-Binding Protein (Leibp) From Arctic Yeast
Organism: Leucosporidium
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2012-02-29 Classification: ANTIFREEZE PROTEIN |
 |
Structural Basis For The Antifreeze Activity Of An Ice-Binding Protein (Leibp) From Arctic Yeast
Organism: Leucosporidium
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2012-02-15 Classification: ANTIFREEZE PROTEIN Ligands: GOL |
 |
Crystal Structure Of A Glutathione S-Transferase From Antarctic Clam Laternula Elliptica
Organism: Laternula elliptica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-02-01 Classification: TRANSFERASE |
 |
Crystal Structure Of A Glutathione-S-Transferase From Antarctic Clam Laternula Elliptica In A Complex With Glutathione
Organism: Laternula elliptica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-02-01 Classification: TRANSFERASE Ligands: GSH |

