Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: DNA |
 |
[2T7-Sio2-Dry] Rhombohedral Tensegrity Triangle Lattice Coated In Silica Tapped Dry And Flash Frozen Without Cryoprotectant
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: DNA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: CELL CYCLE Ligands: A1D6Z |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: CELL CYCLE Ligands: A1D60 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: DNA Ligands: CU |
 |
Cryo-Em Structure Of The Zeaxanthin-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, K3I |
 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, A1L4O |
 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, A1L4O, CL, PC1, PLC, R16, 8K6, D12, C14, D10 |
 |
Cryo-Em Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, CL, PC1, PLC, D12, R16, 8K6, C14 |
 |
Organism: Salmonella enterica
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: HYDROLASE Ligands: MG |
 |
Organism: Salmonella enterica, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: HYDROLASE Ligands: MG |
 |
Organism: Salmonella enterica, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: HYDROLASE Ligands: DGT, MG |
 |
Organism: Salmonella enterica
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: HYDROLASE Ligands: TTP, DGT, MG |
 |
Organism: Shewanella putrefaciens cn-32
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: MG |
 |
Structure Of Photosynthetic Lh1-Rc Complex The Halophilic Nonsulfur Purple Bacterium, Rhodothalassium Salexigens
Organism: Rhodothalassium salexigens dsm 2132
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: PHOTOSYNTHESIS Ligands: HEM, HEC, Z41, PLM, MG, CA, BCL, BPH, U10, LMT, PGV, FE, A1L8Q, CRT |
 |
Glycoside Hydrolase Family 1 Beta-Glucosidase (E318G Mutant) From Streptomyces Griseus (Sophorose Complex)
Organism: Streptomyces griseus subsp. griseus nbrc 13350
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: HYDROLASE Ligands: PGE, CL, BGC |
 |
The X-Ray Structure Of N-Terminal Catalytic Domain Of Thermoplasma Acidophilum Trna Methyltransferase Trm56 (Ta0931) In Complex With 5'-Methylthioadenosine
Organism: Thermoplasma acidophilum dsm 1728
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: MTA |
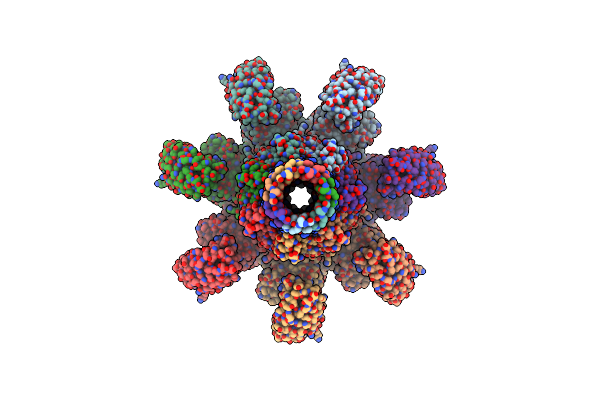 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TOXIN Ligands: CA |
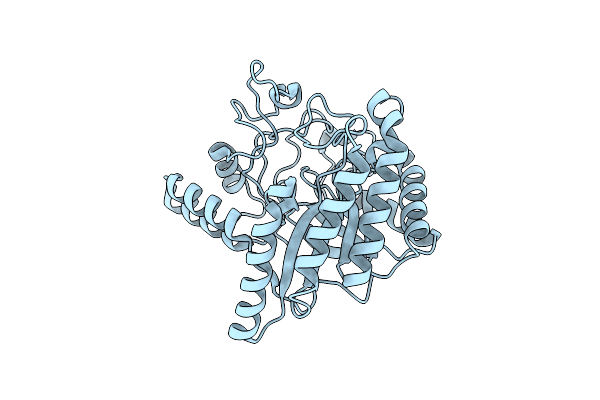 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.36 Å, 1.86 Å Release Date: 2025-03-12 Classification: HYDROLASE |