Search Count: 55
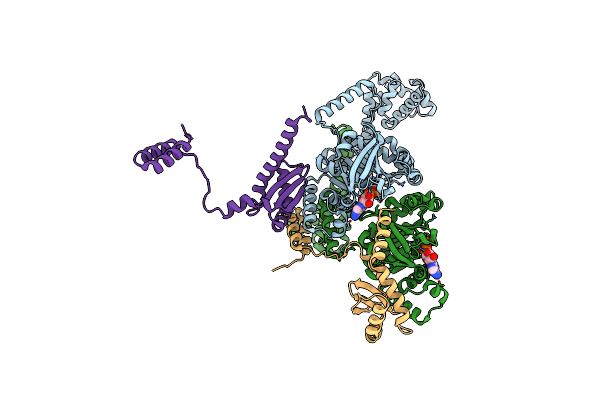 |
Crystal Structure Of The Vibrio Cholerae Replicative Helicase (Vcdnab) In Complex With Its Loader Protein (Vcdcia)
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-02-15 Classification: REPLICATION Ligands: ADP, MG |
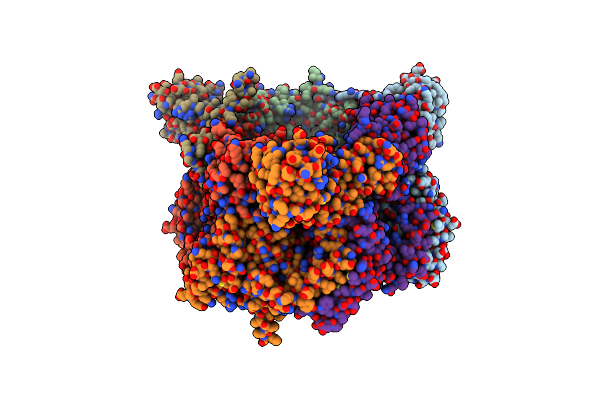 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2022-05-25 Classification: REPLICATION |
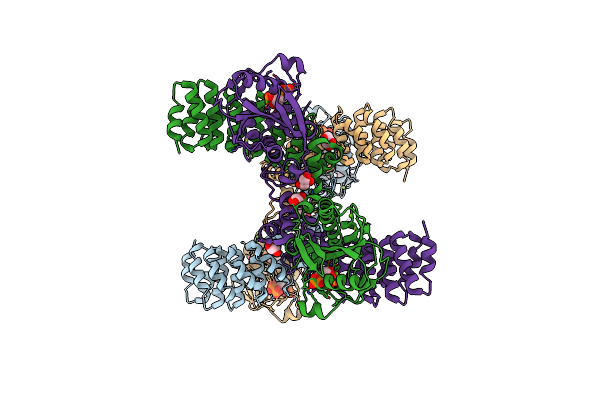 |
Crystal Structure Of Helicobacter Pylori Comf Fused To An Artificial Alpharep Crystallization Helper(Named B2)
Organism: Synthetic construct, Helicobacter pylori (strain atcc 700392 / 26695)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-04-06 Classification: RECOMBINATION Ligands: ZN, PRP, MG, GOL |
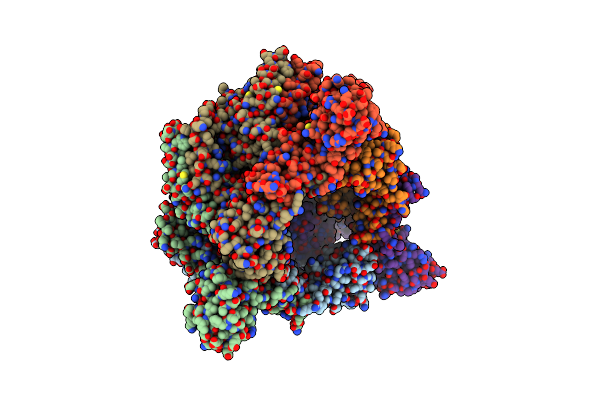 |
Crystal Structure Of The Vibrio Cholerae Replicative Helicase (Dnab) With Gdp-Alf4
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2021-04-28 Classification: REPLICATION Ligands: GDP, MG, ALF |
 |
The Ctd Of Hpdpra, A Dna Binding Winged Helix Domain Which Do Not Bind Dsdna
Organism: Helicobacter pylori
Method: SOLUTION NMR Release Date: 2019-04-24 Classification: DNA BINDING PROTEIN |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2018-01-17 Classification: DNA BINDING PROTEIN Ligands: ACT, GOL, CL |
 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-12-20 Classification: DNA BINDING PROTEIN Ligands: EDO |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2014-02-19 Classification: UNKNOWN FUNCTION |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2014-02-19 Classification: UNKNOWN FUNCTION |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2014-02-12 Classification: TRANSCRIPTION |
 |
Structure Of Pav1-137, A Protein From The Virus Pav1 That Infects Pyrococcus Abyssi.
Organism: Pyrococcus abyssi virus 1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-06-19 Classification: UNKNOWN FUNCTION |
 |
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2012-12-05 Classification: CELL CYCLE Ligands: SO4, CL, EDO |
 |
X-Ray Structure Of Dna Processing Protein A (Dpra) From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-08-29 Classification: DNA BINDING PROTEIN Ligands: SO4 |
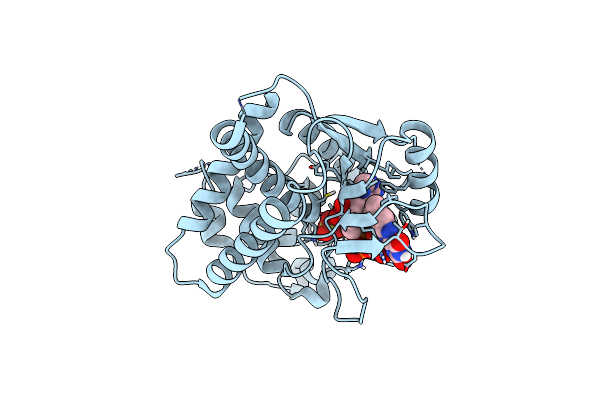 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-05-26 Classification: TRANSFERASE Ligands: SO4, FAD |
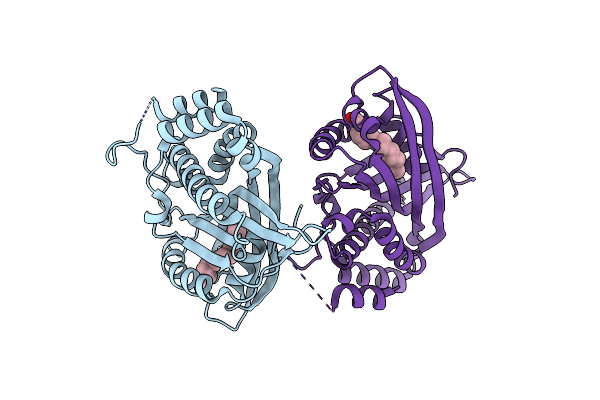 |
Organism: Pectobacterium carotovorum subsp. carotovorum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-12-30 Classification: LIPID BINDING PROTEIN Ligands: PLM |
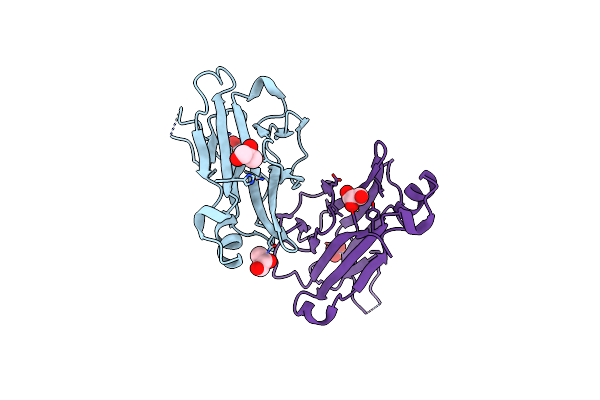 |
Structure Of The Yeast Pml1 Splicing Factor And Its Integration Into The Res Complex
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-09-26 Classification: GENE REGULATION Ligands: GOL, SO4 |
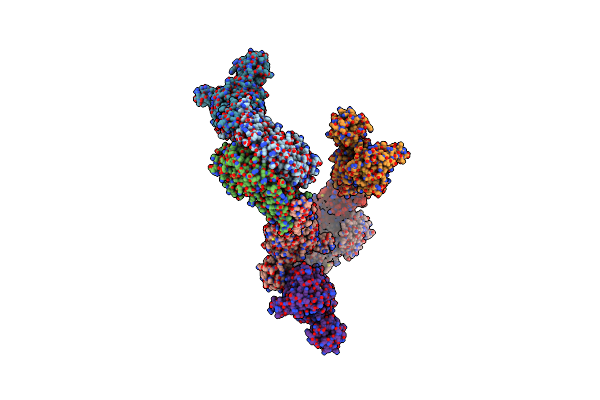 |
Crystal Structure Of An Intact Type Ii Dna Topoisomerase: Insights Into Dna Transfer Mechanisms
Organism: Sulfolobus shibatae
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2008-02-12 Classification: ISOMERASE Ligands: RDC |
 |
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2007-07-31 Classification: HYDROLASE Ligands: GOL, ANP, MG |
 |
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-07-31 Classification: HYDROLASE Ligands: WO4 |
 |
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-07-31 Classification: HYDROLASE Ligands: FE2, ATP |

