Search Count: 38
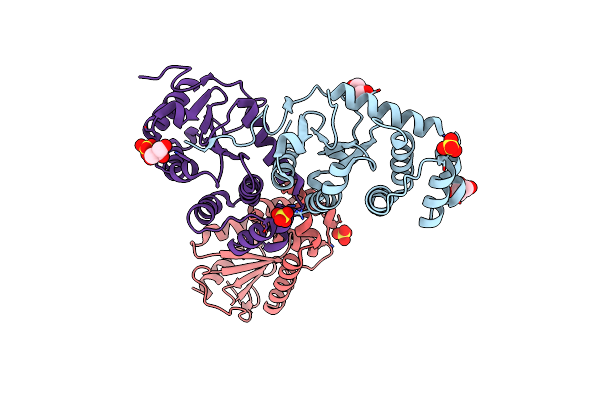 |
Crystal Structure Of Disulfide Bond Oxidoreductase Dsba1 From Legionella Pneumophila
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2013-04-24 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
Crystal Structure Of Mouse Apolipoprotein A-I Binding Protein In Complex With Thymine.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-07-18 Classification: PROTEIN BINDING Ligands: TDR, SO4 |
 |
Crystal Structure Of Mouse Apolipoprotein A-I Binding Protein In Complex With Thymidine
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2012-07-18 Classification: PROTEIN BINDING Ligands: THM |
 |
Crystal Structure Of Mouse Apolipoprotein A-I Binding Protein In Complex With Thymidine 3'-Monophosphate
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2012-07-18 Classification: PROTEIN BINDING Ligands: T3P, SO4 |
 |
Crystal Structure Of Mouse Apolipoprotein A-I Binding Protein In Complex With Theophylline
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-07-18 Classification: PROTEIN BINDING Ligands: SO4, TEP |
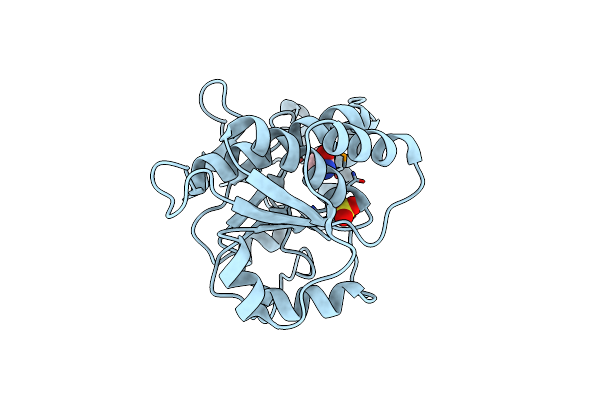 |
Crystal Structure Of Mouse Apolipoprotein A-I Binding Protein In Complex With Nicotinamide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-07-18 Classification: PROTEIN BINDING Ligands: SO4, NCA |
 |
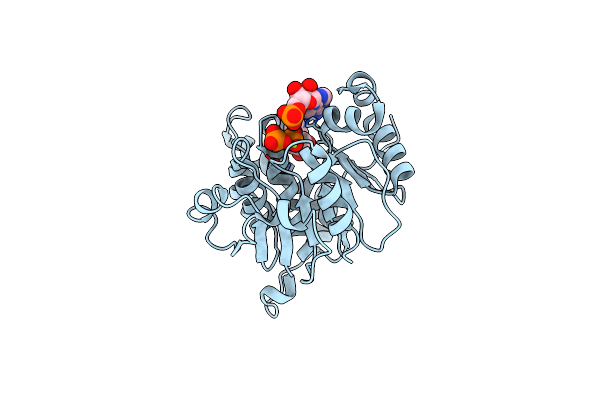
Crystal Structure Of Mouse Apolipoprotein A-I Binding Protein In Complex With Nadp.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-05-09 Classification: PROTEIN BINDING Ligands: NAP |
 |
Crystal Structure Of Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Bacillus Subtilis Co-Crystallized With Atp/Mg2+.
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-07-27 Classification: lyase/lyase substrate Ligands: MG, AMP, PO4 |
 |
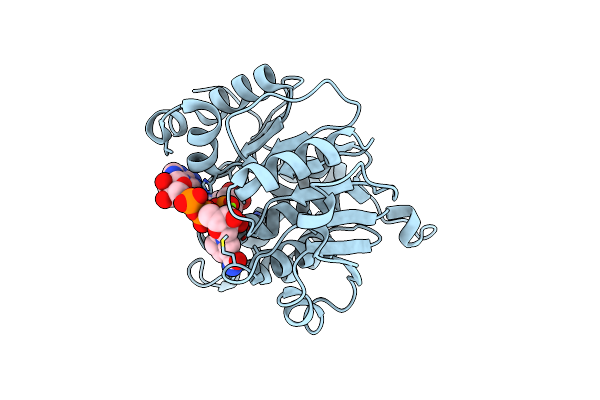
Crystal Structure Of Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Bacillus Subtilis Co-Crystallized With Atp/Mg2+ And Soaked With Nadph
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2011-07-27 Classification: lyase/lyase substrate Ligands: MG, AMP, NPW |
 |
Crystal Structure Of Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Bacillus Subtilis Co-Crystallized With Atp/Mg2+ And Soaked With Nadh
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-07-27 Classification: lyase/lyase substrate Ligands: MG, AMP, NAX |
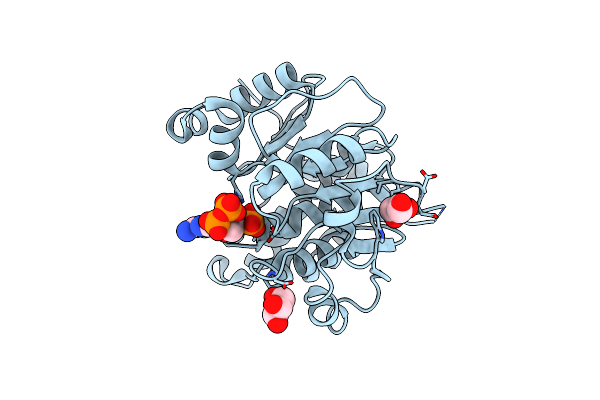 |
Crystal Structure Of Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Bacillus Subtilis Co-Crystallized With Atp/Mg2+ And Soaked With Coa
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-07-27 Classification: lyase/lyase substrate Ligands: COA, GOL |
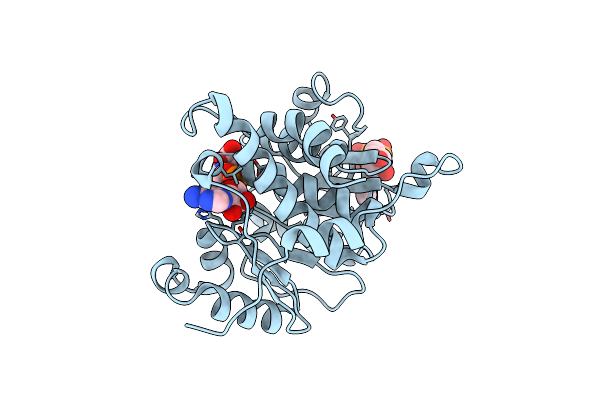 |
Crystal Structure Of Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Bacillus Subtilis Soaked With Adp-Ribose
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-07-27 Classification: lyase/lyase substrate Ligands: MG, APR |
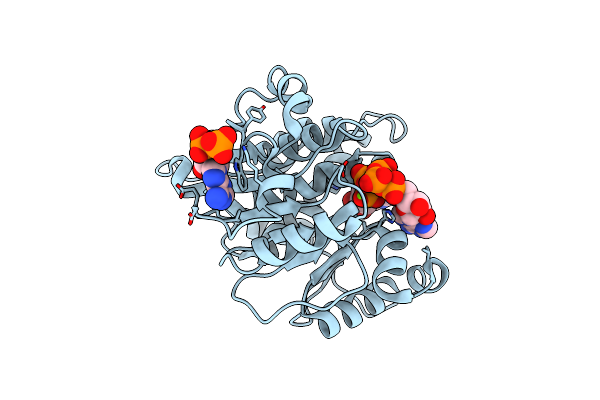 |
Crystal Structure Of Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Bacillus Subtilis Soaked With P1,P5-Di(Adenosine-5') Pentaphosphate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-07-27 Classification: lyase/lyase substrate Ligands: MG, AP5 |
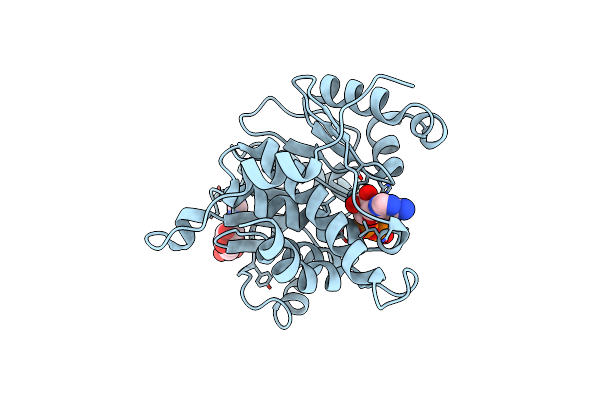 |
Crystal Structure Of Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Bacillus Subtilis In Complex With P1,P6-Di(Adenosine-5') Hexaphosphate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-07-27 Classification: lyase/lyase substrate Ligands: MG, B6P |
 |
Crystal Structure Of Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Bacillus Subtilis In Complex With P1,P3-Di(Adenosine-5') Triphosphate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-07-27 Classification: lyase/lyase substrate Ligands: MG, BA3 |
 |
Crystal Structure Of Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Bacillus Subtilis In Complex With P1,P4-Di(Adenosine-5') Tetraphosphate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-07-27 Classification: lyase/lyase substrate Ligands: MG, B4P, CL |
 |
Crystal Structure Of Tm0922, A Fusion Of A Domain Of Unknown Function And Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Thermotoga Maritima In Complex With Amp
Organism: Thermotoga maritima, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-07-27 Classification: LYASE Ligands: K, AMP |
 |
Crystal Structure Of Tm0922, A Fusion Of A Domain Of Unknown Function And Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Thermotoga Maritima In Complex With Adp
Organism: Thermotoga maritima, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2011-07-27 Classification: LYASE Ligands: K, MG, ADP, GOL |
 |
Crystal Structure Of Tm0922, A Fusion Of A Domain Of Unknown Function And Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Thermotoga Maritima In Complex With Atp
Organism: Thermotoga maritima, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-07-27 Classification: LYASE Ligands: K, MG, ATP, GOL |
 |
Crystal Structure Of Tm0922, A Fusion Of A Domain Of Unknown Function And Adp/Atp-Dependent Nad(P)H-Hydrate Dehydratase From Thermotoga Maritima In Complex With P1,P5-Di(Adenosine-5') Pentaphosphate
Organism: Thermotoga maritima, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-07-27 Classification: LYASE Ligands: K, AP5, GOL |

