Search Count: 17
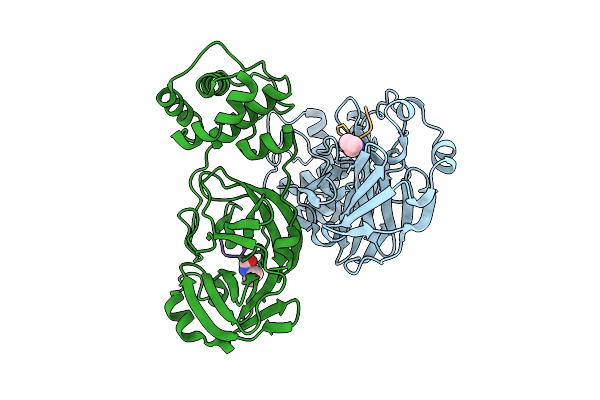 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBOTOR Ligands: A1L7M |
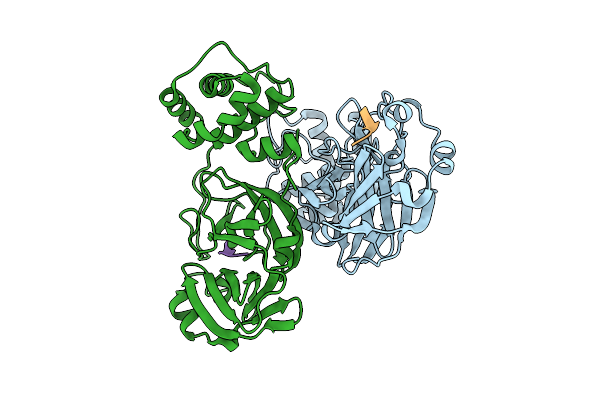 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
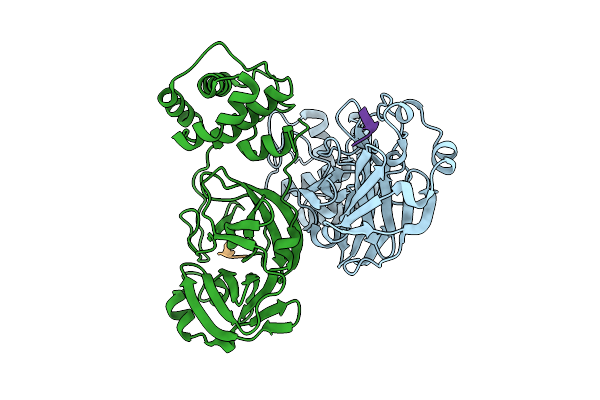 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
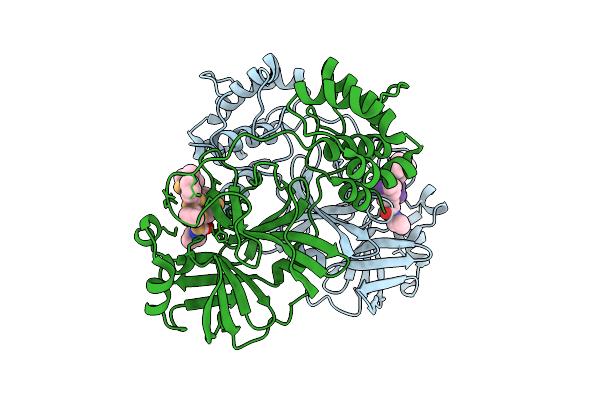 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR Ligands: A1L7M |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR Ligands: A1L7M |
 |
Identification Of Small-Molecule Binding Sites Of A Ubiquitin-Conjugating Enzyme-Ube2T Through Fragment-Based Screening
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-02-28 Classification: LIGASE Ligands: V23, EDO |
 |
Identification Of Small-Molecule Binding Sites Of A Ubiquitin-Conjugating Enzyme-Ube2T Through Fragment-Based Screening
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-02-28 Classification: LIGASE Ligands: V2R |
 |
Identification And Characterization Of Inhibitors Covalently Modifying Catalytic Cysteine Of Ube2T And Blocking Ubiquitin Transfer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-11-29 Classification: LIGASE/INHIBITOR Ligands: V6C, EDO |
 |
Identification And Characterization Of Inhibitors Covalently Modifying Catalytic Cysteine Of Ube2T And Blocking Ubiquitin Transfer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-11-29 Classification: LIGASE/INHIBITOR Ligands: V5L, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.26 Å Release Date: 2019-06-26 Classification: TRANSFERASE Ligands: AFU |
 |
Organism: Streptococcus pneumoniae ga47502
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-05 Classification: ANTIMICROBIAL PROTEIN Ligands: 54B |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: MES |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
Structure Of A Deazaflavin-Dependent Nitroreductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42 |
 |
Structure Of A Deazaflavin-Dependent Reductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42, SO4 |

